Abstract
Marine cyanobacteria of the genus Prochlorococcus represent numerically dominant photoautotrophs residing throughout the euphotic zones in the open oceans and are major contributors to the global carbon cycle. Prochlorococcus has remained a genetically intractable bacterium due to slow growth rates and low transformation efficiencies using standard techniques. Our recent successes in cloning and genetically engineering the AT-rich, 1.1 Mb Mycoplasma mycoides genome in yeast encouraged us to explore similar methods with Prochlorococcus. Prochlorococcus MED4 has an AT-rich genome, with a GC content of 30.8%, similar to that of Saccharomyces cerevisiae (38%), and contains abundant yeast replication origin consensus sites (ACS) evenly distributed around its 1.66 Mb genome. Unlike Mycoplasma cells, which use the UGA codon for tryptophane, Prochlorococcus uses the standard genetic code. Despite this, we observed no toxic effects of several partial and 15 whole Prochlorococcus MED4 genome clones in S. cerevisiae. Sequencing of a Prochlorococcus genome purified from yeast identified 14 single base pair missense mutations, one frameshift, one single base substitution to a stop codon and one dinucleotide transversion compared to the donor genomic DNA. We thus provide evidence of transformation, replication and maintenance of this 1.66 Mb intact bacterial genome in S. cerevisiae.
INTRODUCTION
Cyanobacteria of the genus Prochlorococcus are estimated to be the most abundant photosynthetic organisms on the planet; they are commonly distributed throughout the euphotic zone at cell concentrations ranging from 104 to 105 cells/ml and are a major contributor to the global carbon cycle. Prochlorococcus species have adapted to nutrient-poor environments and can thrive in high-light as well as low-light conditions. Although only 0.5–1 µm in diameter, it is estimated that Prochlorococcus populations constitute 30–60% of total chlorophyll biomass between latitudes 40°N and 40°S (1–3). Prochlorococcus marinus MED4 has been described as a high-light adapted ecotype (4). It contains the smallest genome of any known oxygenic phototroph [1 657 990 bp encoding 1716 annotated genes].
Genetic tools for manipulation of Prochlorococcus are largely lacking. There are no known natural plasmids and thus far transformation methods such as Polyethylene Glycol (PEG)-mediated transformation or electroporation have not been developed. Lateral gene transfer among Prochlorococcus strains has been observed frequently when mediated by cyanophages and appears to be an important evolutionary mechanism (5). There is one report of insertion of transposable elements into the Prochlorococcus MIT9313 genome by plasmid conjugation; however, Prochlorococcus strains have by and large remained genetically intractable (6–9).
The MED4 strain grows in artificial sea water (Pro99 medium) with a minimal doubling time of 24 h under bright light. Consequently, colonies derived from individual cells develop only after several weeks. The native Prochlorococcus MED4 strain was sequenced by the Joint Genome Institute in August 2003 and deposited as an axenic culture at the National Center for Marine Algae and Microbiota (NCMA, formerly the CCMP) in February 2004. Since then MED4 cultures have not been cryopreserved, but continuously passaged in Pro99 medium (personal communication with NCMA/CCMP curator), thus the MED4 strain has not been maintained in a way that guarantees clonal purity.
Our recent achievements in cloning Mycoplasma genomes into Saccharomyces cerevisiae by joining them to a yeast vector containing a centromere (CEN) and a yeast selectable marker, our ability to then genetically engineer the cloned genome using yeast genetic tools and finally our ability to transplant the engineered genome back into a recipient Mycoplasma cell to bring the engineered genome back to life (10–15) have led us to attempt a similar approach with Prochlorococcus. We describe in this article the successful cloning of the entire P. marinus MED4 genome in yeast and detailed sequence analysis of the bacterial circular chromosome purified from yeast. This success has depended primarily on two properties of the Prochlorococcus genome, AT richness, which results in a large number of consensus yeast replication origins, and lack of toxic gene products, presumably due to the differences in expression mechanisms between yeast and bacteria.
MATERIALS AND METHODS
Yeast and Prochlorococcus culture conditions
Yeast strains W303-1A (MATa leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 ybp1-1) and VL6-48 (MATα, his3-Δ1, trp1-Δ1, ura3-52, lys2, ade2-101, met14) were described previously (14,16). Prochlorococcus marinus MED4 (CCMP1986) was continuously grown in Pro99 medium at 18 ± 1°C on a 14:10 light:dark cycle at 20 µmol Q m−2 s−1 from cool white, fluorescent bulbs. Approximately every 2 weeks, 3 ml of culture was diluted into 25 ml of fresh medium to maintain the strains. Growth of Prochlorococcus cultures was monitored by fluorometric detection of bulk chlorophyll autofluorescence (Turner Design Fluorometer 10-AU, excitation, 436 nm; absorbance, 680 nm). Cell concentrations of Prochlorococcus cultures were determined by flow cytometry using SYBR Green I fluorescence in a BD FACS Aria-II (488 nm; emission filter at 512 nm).
Isolation of Prochlorococcus genomic DNA for yeast transformation and sequencing
MED4 cells were harvested from a 30-ml culture grown to a bulk chlorophyll autofluorescence of 50 Relative Fluorescence Units (RFUs) by centrifugation, washed twice with 10 mM Tris, 0.5 M sucrose pH 6.5 buffer, resuspended in 500 µl of buffer, equilibrated for 5 min in a 50°C water bath and combined with 500 µl pre-melted 2.2% low melting point (LMP) agarose at 50°C. An equivalent of 1.5 × 108 cells was divided into 10 plugs, resulting in ∼1.5 × 107 cells per plug. Intact genomes were isolated in agarose plugs according to the clamped homogeneous electric fields (CHEF)-DR III Manual (BioRad). After additional washes with 0.1× wash buffer (2 mM Tris, 5 mM EDTA, pH 8) and NEB buffer 2, plugs were digested with SgrDI or RsrII at 37°C overnight (80 U enzyme per plug). Prior to transformation, plugs were melted in 1× TE at 65°C for 10 min and treated with β-agarase (NEB) at 42°C overnight. Concentrations of melted plug DNA were determined to be 20–25 ng/µl by gel electrophoresis of sheared plug DNA and comparison to quantitative ladders. For transformation, 40 µl genomic DNA (equivalent to 0.8–1 µg) was used. For sequencing of wild-type MED4 genomic DNA, plugs were phenol-extracted after melting and agarase treatment.
Preparation of vector DNA for transformation
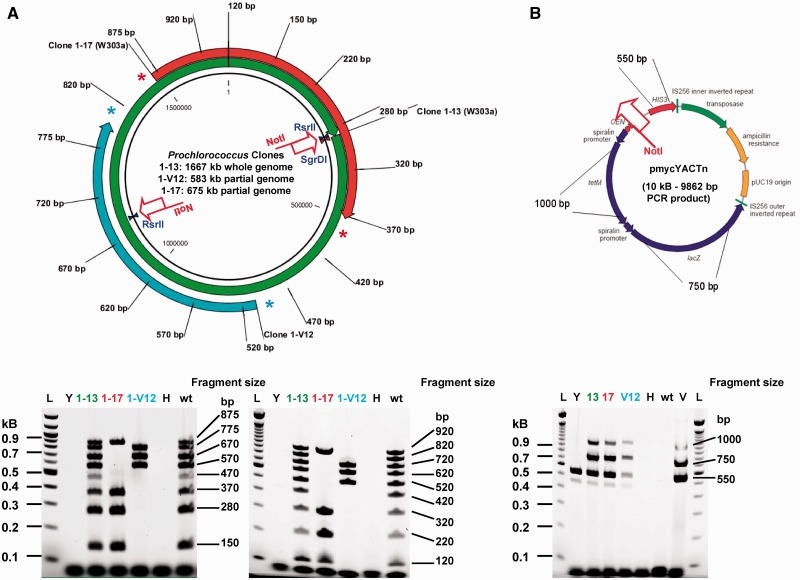
The 10-kb trishuttle pmycYACTn vector (GU593054) contains a yeast centromere (CEN6), a yeast selectable marker (HIS3) and the yeast replication origin (ARSH4) (14). We amplified this with primers that contained 20 bp of sequence flanking the replication origin (so as to exclude it from the PCR product) and 60 bp of sequence to either side of the unique genomic SgrDI restriction site. The PCR product was 9862 bp long; 10 ng was used as template in a 100 -µl reaction with ExTaq (TaKaRa) at final concentrations of 5 U DNA polymerase, 2 mM MgCl2, 0.2 µM each primer and 0.2 mM of each dNTP. Cycling conditions were as follows: 94°C for 3 min; followed by five cycles of 94°C for 30 s, 50°C for 30 s and 68°C for 8 min; followed by 30 cycles of 94°C for 30 s, 62°C for 30 s and 68°C for 8 min, with a final extension of 68°C for 5 min. Each PCR vector product was gel purified on 0.8% LMP agarose gels using β-agarase, phenol extraction and ethanol precipitation to remove remaining background circular vector template. Later experiments included transformations with vector DNA products treated with DpnI prior to transformation to digest any remaining GATC-methylated circular vector DNA derived from dam+ Escherichia coli stocks of plasmid pmycYACTn. In this case, PCRs were first digested with 20 U DpnI for 1 h prior to gel purification. For insertion at the single-cut site SgrDI [nt 318371], primers with the following sequences were used (20 bp homology to vector template in bold): TCTAAGTCTCTAAAGTCACTAAAGGTTGTTCCCCCTGCACCAATACTTTTTTTTAAAACGTTCCATCATTAAAAGATACG and CTTTTAGAGAGGCTAGAACAATTAATAAAAATGAAATAAAAAGACTTAGAAGAGCTGTCGAGCAAGATAAAAGGTAGTAT. For insertion at RsrII sites, pmycYACTn was amplified using primers ATCCAACTATTGTCCAAGTCTGATAAGTTCTTGACTGCTGCCCAACCCATGTGCCAGTCGTTCCATCATTAAAAGATACG and AATCTTATTGGTGTCAACTTAATCAATTACCAAAACAAGAGTGGGCAGAATATTTTGACGAGCAAGATAAAAGGTAGTAT for RsrII site 1 [nt 311110] and primers CACATGCTCCAAGGCTTTTAAGTTTTAGGATTGATCCAGAACTTATAGGTACTGTTATCGTTCCATCATTAAAAGATACG and CTTCAATATCTATTTTTGTATTAGTTCTTTCAGTAATTCCTTTAATAGTTCTCCCTCCCGAGCAAGATAAAAGGTAGTAT for RsrII site 2 [nt 1138584].
Yeast transformation and PCR analysis of yeast clones
Yeast transformations were performed by a spheroplast PEG transformation protocol as described (16) except cultures were grown to OD600 1.5. Per 1 µg genomic DNA isolated from agarose plugs (1 µg=1 fmol Prochlorococcus genome), a 30× molar amount of linear vector PCR product (200 ng=30 fmol vector) was added for transformations of whole genomes in a final end volume of 40 µl. Transformants were plated and incubated for 2–3 days at 30°C on selective plates (lacking histidine). Colonies were picked and repatched onto fresh minus histidine plates to avoid false-positive PCR results due to high amounts of genomic DNA on the original plates. DNA was extracted from ∼107 repatched yeast cells suspended in 250 µl P1 buffer as described previously (17,18). The DNA was screened by PCR amplification of the vector insertion site. Primers were designed to give a 500-bp product at the 5′-vector junction. The 3′-vector junction was confirmed by primers yielding a 1000-bp product (Supplementary Table S8, Supplementary Figure S2). Clones were confirmed for completeness by multiplex PCR. Additionally, three amplicons were designed to test for the HIS3, lacZ and TetM sequences of the yeast vector (multiplex primers are listed in Supplementary Tables S9–S11).
Preparation and sizing of Prochlorococcus genomes from yeast
Total DNA from individual yeast clones containing Prochlorococcus genomes was isolated in agarose plugs as described (11,17). Plugs were first digested with AscI, FseI, MreI and SfiI. These enzymes cleave yeast chromosomes but do not have recognition sites in the Prochlorococcus genome. To remove linear yeast chromosomal DNA fragments, plugs were pre-electrophoresed at constant voltage (5–6V/cm) for 5h at 4°C. Plugs were extracted from the first gel and then digested at the single-cut NotI site (only present in the vector) or the two RsrII sites for 4h, and sized by pulsed-field electrophoresis. Genomic DNA for sequencing of MED4 clone 2–19 was derived from 10 melted and phenol-extracted yeast plugs after digestion of yeast DNA and purification by electrophoresis as described above.
Sequence analysis of the cloned Prochlorococcus genome and of the native genome
Both genomes were sequenced by 454 FLX pyrosequencing and Illumina Solexa sequencing at the Joint Technology Center, J. Craig Venter Institute, Rockville, MD. Approximately 5 µg of genomic DNA was used for library construction. Sequence reads from each genome were directly mapped to the 2003 reference genome (BX548174) using CLC Bio Genomics Workbench software version 5.1. To exclude any large insertions or gaps in the sequencing results of the MED4 genomes, de novo assembly of the native genomic DNA reads and the yeast clone reads from both platforms was carried out using MUMmer 3.23 (19). Single nucleotide polymorphism (SNP) and DIP (deletion/insertion polymorphism) detection with both software packages resulted in the same variants described below. For further SNP analysis, primers were designed to amplify open reading frames (ORFs) of genes PMM0039, PMM0214, PMM0844, PMM1600 and PMM1294 from genomic DNA isolated from yeast clone agarose plugs. PCR products were purified using PureLink® PCR Purification Kits (Invitrogen™) and screening of these candidate genes from 15 whole-genome clones in yeast was subsequently carried out by Sanger sequencing using either the 5′- or 3′-primer used for amplification.
Yeast clone growth rates and yeast host mutation complementation experiments
Doubling times were determined by measuring the OD600 of exponentially growing cultures in media lacking histidine at 1-h intervals for at least three generations. Growth rate measurements were repeated three times for each culture. Standard deviations were calculated from the average doubling time differences obtained from each day. To compare growth of whole Prochlorococcus genome clones in yeast, clones 1–13 (first transformation), 2–2 and 2–19 (second transformation) were grown in parallel to a W303-1A strain containing only the cloning vector pmycYACTn (Supplementary Chart S1). The same four yeast clones were spotted as equal cell amounts on -HIS, -HIS-LYS, -HIS-URA, -HIS-LEU and -HIS-TRP drop-out plates to determine whether the bacterial genome would complement the yeast host marker mutations of strain W303-1A (Supplementary Figure S6).
RESULTS
Analysis of putative yeast replication origins in Prochlorococcus MED4
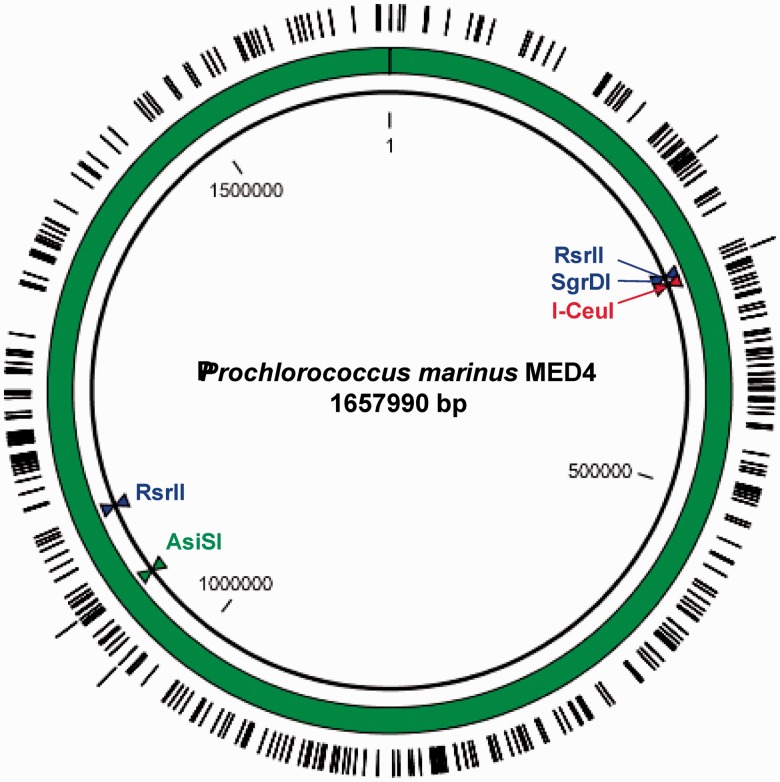
Replication origins in S. cerevisiae are termed autonomously replicating sequence (ARS), and contain ARS consensus sequences (ACS) that are essential for binding of origin replication complexes. An 11-bp ACS consensus sequence (A/T)TTTA(T/C)(A/G)TTT(A/T) has been proposed (20). Additionally, a 17-bp consensus sequence or extended ACS (EACS) was determined by microarray analysis which combined evolutionary conserved ARS sequences as (A/T)(A/T)(A/T)(A/T)TTTA(T/C)(A/G) TTT(A/T)GTT (21–23). Although yeast replication has been studied extensively, it is difficult to fully define a functional replication origin. While a functional ARS generally contains an exact match, or is very similar to the 11-bp consensus sites, and contains at least one A/T-rich region of DNA functioning as an unwinding element, specific binding of replication complexes relies on varying neighboring sequence modules (24,25). We expected that the AT-rich (30.8% GC-content) Prochlorococcus genome would contain a high number of ACS sites. Using motif searches, we identified 328 11-bp ACS and 4 17-bp ACS sites in the genome. The maximum gap between two sites was 34.5 kb (Figure 1). We consider that this type of motif search resulting in an even distribution of ACS sites might be used as a proxy to determine whether a bacterial chromosome will be maintained in yeast. Analysis of previously cloned genomes of Mycoplasma genitalium, Mycoplasma pneumoniae M129, Mycoplasma mycoides (14) and Acholeplasma laidlawii PG-8A (18) showed similar distributions of ACS sites (Supplementary Figure S1). A more recent study showed that fragments with higher GC-content (55%) derived from Synechococcus elongatus PCC7942 and lacking ACS sites could be stably maintained as a 454-kb plasmid in yeast after ARS sequences were inserted at intervals of <150 kb (26).
Figure 1.
Prochlorococcus MED4 genome with single restriction sites SgrDI, AsiSI and I-CeuI. RsrII cuts the genome in half at 311110 and 1138584 bp. The 11 - or 17-bp ARS consensus sequences are evenly distributed along the AT-rich (30.8% GC-content) genome (328 instances of the 11-bp ACS are marked on the inner ring, four instances of the 17 bp site are on the outer ring). The largest segments not containing an ACS are 34.1 and 34.5 kb in size.
Cloning of Prochlorococcus MED4 into yeast
We initially attempted cloning SgrDI linearized whole-genome fragments into yeast using gel-purified genomic DNA. Agarose plugs digested with SgrDI were run on LMP agarose CHEF gels and 1.66 Mb bands were isolated for transformation. Various genomic DNA:vector ratios were tested and although up to 19 colonies were obtained per transformation reaction none yielded Prochlorococcus DNA clones (Supplementary Table S4).
Subsequent transformations used genomic DNA digested with RsrII or SgrDI while in the plug. The cleaved DNA was then recovered from the plug and used directly for transformations. This yielded RsrII half genomes (clone 1–17, 675 kb and clone 1–V12, 580 kb) and whole-genome clones derived from SgrDI digests. Additionally, these only appeared in our screen for transformations using a genomic DNA:vector molar ratio of 1:30, while 1:3 and 1:10 ratios yielded no Prochlorococcus clones (Figure 2, Supplementary Table S5).
Figure 2.
Analysis of Prochlorococcus MED4 clones by multiplex PCR. (A) Primer pairs annealing to sites distributed along the genome every 100 kb show evidence for two partial clones (clone 1–17 in yeast strain W303-1 A, ∼675 kb in size and clone 1–V12 in yeast strain VL6-48, 583 kb in size) and one whole genome (clone 1–13, 1667 kb in size) isolated from S. cerevisiae strain W303-1 A as compared to wild-type Prochlorococcus genomic DNA (wt) multiplex patterns. (B) Presence of the cloning vector only was confirmed by screening with three amplicons specific for the vector sequence (HIS3: 550 bp, lacZ: 750 bp and TetM: 1000 bp). All three clones were positive for these markers compared to negative controls (H = H2O, wt = Prochlorococcus genomic DNA). Only the endogenous his3-11,15 allele is also amplified by the HIS3 primer pair as expected in wt W303-1 A yeast genomic DNA (Y); pmycYACTn vector only (V); 100-bp ladder, Invitrogen (L).
All yeast clones that showed evidence of one or more multiplex amplicons specific for MED4 were cultured to saturation in selective medium. DNA was isolated in agarose plugs and examined for size by restriction digestion followed by CHEF gel electrophoresis (Figure 3). We presumed some clones recombined into smaller circles due to fragmented genomic DNA derived from the Prochlorococcus agarose plugs (Supplementary Figures S2 and S3).
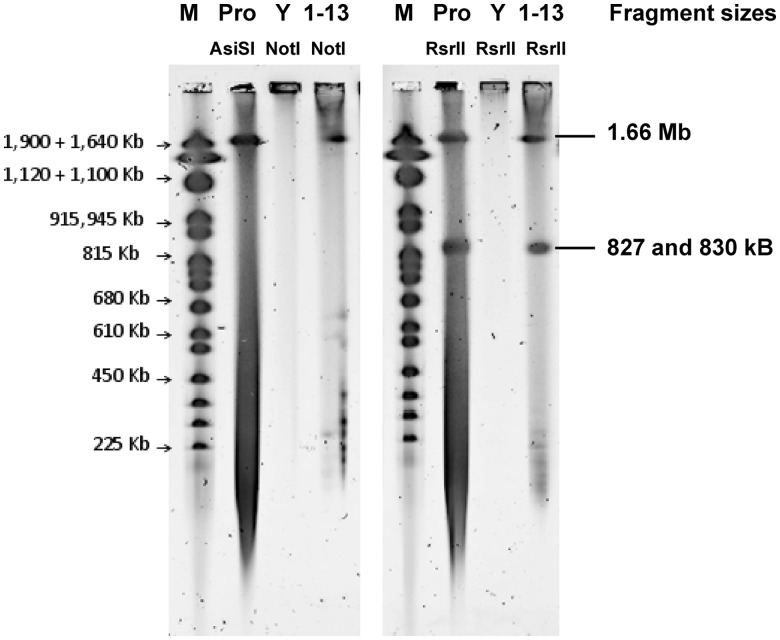
Figure 3.
CHEF gel analysis of a Prochlorococcus MED4 whole genome (clone 1–13) in W303-1 A. Agarose plugs were prepared from wild-type Prochlorococcus MED4 cells (Pro), wild-type W303-1 A yeast cells (Y) and W303-1 A Prochlorococcus MED4 clone 1–13 cells and treated to isolate genomic DNA. Plugs containing only wt Prochlorococcus genomic DNA were digested with AsiSI for whole linear genomes (Pro, lane 2) or RsrII for half-genome fragments (Pro, lane 6). Plugs containing wt W303-1 A yeast cells or clone 1–13 yeast cells were digested with AscI, FseI, MreI and SfiI. These enzymes cleave yeast chromosomes but do not have recognition sites in the Prochlorococcus MED4 genome. Electrophoresis of plugs at constant voltage (100 V) for 90 min in a 1% Tris Acetate EDTA (TAE) gel removed linear yeast chromosomal DNA, while large circular Prochlorococcus DNA molecules remain topologically trapped in the plugs. Clone 1–13 or wt W303-1 A plugs were then digested with the single-cutter NotI (only present in the cloning vector of clone 1–13, lane 4) or RsrII (1–13, lane 8). RsrII cuts the Prochlorococcus MED4 genome twice into 827 and 830 kb fragments. Yeast chromosome marker (M).
A third transformation experiment yielded a number of partial clones containing a range of multiplex amplicons as well as two more whole-genome clones (Supplementary Figures S3 and S4). Transformations with DpnI-digested vector product seemed to significantly reduce the background (Supplementary Table S6).
A fourth transformation experiment yielded a higher cloning efficiency resulting in 12 whole-genome clones out of 16 yeast colonies. All transformations using a DpnI-treated vector resulted in whole-genome clones (Supplementary Table S7, Supplementary Figure S5). Some background colonies can arise from recircularization of the linear vector PCR product itself or of the linear vector possibly inserting into yeast genomic DNA to generate histidine prototrophy. Though each transformation was carried out with a 1:30 gDNA:vector ratio, it is difficult to predict the number of intact genomes per plug that contribute to a higher yield of whole clones in yeast. As such, these ratios are only a general approximation to follow when cloning genomic DNA.
Sequence analysis of the native Prochlorococcus MED4 genome
The native Prochlorococcus MED4 strain was sequenced by the Joint Genome Institute in August 2003 and was deposited as an axenic culture at the NCMA in February 2004. Since then, MED4 cultures have not been cryopreserved, but have been continuously passaged in Pro99 medium (personal communication with NCMA/CCMP curator). We sequenced the native strain obtained from the culture collection to determine any spontaneous mutations that may have occurred and been fixed at high frequency in the population over the last 8 years. Of 108 496 sequencing reads obtained by 454 sequencing, 108 310 reads mapped to cover the 1 657 990 bp native GenBank reference sequence (BX548174). Of the subsequent 35 672 668 Illumina reads, 35 530 402 matched to the GenBank reference. Combined with de novo assembly of all reads, we identified 27 mutations, 24 of which were single base pair variants, to the Genbank reference sequence (Supplementary Table S1).
A putative sulfate transporter gene (PMM0214) showed four missense mutations. Other non-synonymous SNPs were found in the transcription factor NtcA, the (ppGpp)ase SpoT, the Photosystem II gene PsbC, cytochrome cM, a Na+/H+ antiporter and molecular chaperones DnaJ and DnaK. Osburne et al. (27) resequenced a wild-type Prochlorococcus MED4 strain in 2008. The Illumina sequencing results of that study showed 17 variants compared to the 2003 reference sequence. Six of these were identical ones found in our sequencing results (Supplementary Table S1). The native MED4 strain deposited at the NCMA/CCMP in 2004 and serial passaging over 8 years has induced a remarkably small number of variations. We assume that the native cultures analyzed in the above sequencing projects are a mix of clones and therefore one cannot attribute any single or all identified mutations to one clone.
Comparison of the yeast clone to the native sequence
Sequencing data from the Prochlorococcus genome cloned in yeast (clone 2–19) showed a high yeast genome background even after gel purification of the linear yeast chromosomes from the circular bacterial genome topologically trapped in genomic DNA agarose plugs. Of the 792 302 total reads obtained by 454 sequencing, 288 853 reads mapped to the bacterial genome with an average coverage of 50× along the complete 1 657 990 bp GenBank reference. Sixty-three percent of unmapped reads can be attributed to the high yeast genomic DNA background (specifically 492 710 reads mapped to S. cerevisiae chromosomes). Of the remaining 10 739 reads, 1927 mapped to the cloning vector sequence inserted at the SgrDI site as designed. Illumina sequencing showed 3 350 748 out of 40 370 844 reads matched to complete the Prochlorococcus GenBank reference genome with an average 250× coverage.
The inserted vector sequence was found to contain 40 single base pair alterations, presumably introduced during PCR amplification rather than during transformation into yeast. Among SNPs identified, the ampicillin resistance gene showed one glutamine to glycine mutation, lacZ had three amino acid substitutions and the tetracycline resistance gene had 12 amino acid substitutions (Supplementary Table S2).
The cloned genome contained 27 mutational changes (24 were SNPs) compared to our sequence of the native Prochlorococcus genome. Differences are summarized in Supplementary Table S3. Nine SNPs represent positions at which the yeast clone is identical to the original 2003 reference genome sequence. Our yeast MED4 clone 2–19 represents a single genome from the MED4 culture at the time of harvest. Therefore, these nine SNPs most likely represent sequence heterogeneity at these positions within the culture, rather than reverse mutations to the original 2003 reference genome. Sixteen further single base pair variations were identified in the 2–19 yeast clone. Six represent silent mutations or were found in intergenic regions, eight SNPs cause amino acid substitutions, one is an ORF frameshift mutation and one is a mutation to a stop codon which causes an ORF truncation. An additional dinucleotide insertion (switching threonine to valine) and one amino acid change induced by a SNP occurred in the previously described sulfate transporter gene (PMM0214), which was found to be mutated at four different sites in the native genome. Another transporter gene (PMM1600, a putative Na+/H+ antiporter), mutated in the native genome analysis (two SNPs, see Supplementary Table S1), was mutated in the yeast clone also. Further missense point mutations were identified in a possible type I restriction–modification enzyme, a putative modulator of DNA gyrase (TldD), a seryl-tRNA synthetase (SerS) and in the RNA polymerase β-subunit (RpoB).
Analysis of gene mutations in other yeast clones
Each of the cloning experiments described above was derived from a separate genomic DNA preparation of the native culture. We obtained 1 whole-genome clone in our first successful transformation (clone 1–13), 2 more in our second attempt (clones 2–2 and 2–19) and 12 more in our third attempt (clones 3–2, 3–3, 3–7 to 3–16). These clones were screened for some of the SNPs identified in the whole-genome sequencing of yeast clone 2–19. Five candidate genes were chosen that showed mutations in clone 2–19 compared to the native sequencing data (highlighted in Supplementary Table S3). PCR amplification from genomic DNA prepared from the native Prochlorococcus culture and the 15 different MED4 whole-genome yeast clones was carried out for genes PMM0039, PMM0214, PMM0844, PMM1600 and PMM1294 (Table 1, Supplementary Figure S5). Sanger sequencing of the respective PCR products from the native culture detected the same genotype as for the initial native bacterial culture DNA preparation identified by 454 and Illumina sequencing. The five PCR products from yeast clone 2–19 also showed the same mutations as the whole-genome sequencing data (compare Supplementary Table S3 and Table 1). The remaining yeast clones (1–13, 2–2, 3–2, 3–3, 3–7 to 3–16) contained both versions of either the original native sequence or the mutations identified in clone 2–19 (Table 1). Therefore, the identified insertions, transversions or SNPs do not represent loss of function mutations upon cloning into the yeast host cells.
Table 1.
Analysis of five candidate genes of all MED4 whole genome yeast clones to the native references
| Gene in MED4 genome | PMM0039 | PMM0214 | PMM0844 | PMM1600 | PMM1294 |
|---|---|---|---|---|---|
| Position in reference sequence | 37452-3 | 208537-8 | 803532 | 1529073 | 1246342 |
| Bacterial genomes | – | ||||
| MED4 reference (BX548174) | – | AC | C | C | C |
| MED4 JCVI (fully sequenced de novo) | AC | C | C | T | |
| Yeast (MED4 whole-genome clones) | |||||
| Clone 1–13 | – | AC | C | C | C |
| Clone 2–2 | – | GT | C | C | C |
| Clone 2–19 (fully sequenced de novo) | T insertion | GT | A | T | C |
| Clone 3–2 | T insertion | GT | A | T | C |
| Clone 3–3 | T insertion | GT | A | T | C |
| Clone 3–7 | T insertion | GT | C | T | C |
| Clone 3–8 | T insertion | GT | A | T | C |
| Clone 3–9 | T insertion | GT | C | T | C |
| Clone 3–10 | T insertion | GT | A | T | C |
| Clone 3–11 | – | GT | A | C | C |
| Clone 3–12 | T insertion | GT | A | T | C |
| Clone 3–13 | T insertion | GT | A | T | C |
| Clone 3–14 | – | GT | A | C | C |
| Clone 3–15 | – | GT | A | C | C |
| Clone 3–16 | GT | C | C | C |
Comparison of one insertion, one transversion and three SNPs in five candidate genes between 15 MED4 yeast clones to the MED4 native genomes. Individual mutations from PCR product sequences of PMM0039, PMM0214, PMM0844, PMM1600 and PMM1294, each derived from 15 different whole-genome Prochlorococcus MED4 clones in yeast strain W303-1A (Supplementary Figure S5) are compared to PCR product sequences from native MED4 genomic DNA (JCVI) by Sanger sequencing. The genotype of the original 2003 Genbank reference genome (BX548174) is shown on top.
Our results are consistent with the existence of a mixture of clones in the original MED4 culture. Prior analysis of yeast clone sequences from native M. mycoides (24% GC-content) or M. genitalium (32% GC-content) genome assemblies transformed into yeast using the same protocol showed no mutations compared to the donor DNA. Furthermore, the M. mycoides genome isolated out of yeast was transplanted and aside from changes that were engineered in yeast, it contained the identical sequence as the molecule that was initially cloned into yeast (12–13).
Growth rate analysis of yeast clones
The three whole-genome Prochlorococcus yeast clones 1–13, 2–2 and 2–19 were stored frozen in 25% glycerol at −80°C. They were repatched on selective plates (lacking histidine), passaged in liquid medium (lacking histidine) and analyzed by multiplex PCR and CHEF gels. The banding patterns were the same for all three clones (Supplementary Figures S3 and S4). Growth rates of the whole-genome clones were compared in three separate experiments to the wild-type yeast W303-1 A strain carrying only the circular cloning vector. Doubling times of the three clones compared to the plasmid clone were nearly identical (Supplementary Chart S1). Additionally, Prochlorococcus genomes are stably maintained in cultures grown not only in liquid media (lacking histidine) to late stationary phase but also were detectable by multiplex or pulsed-field gel analysis from cells grown in non-selective yeast extract peptone dextrose complex medium (YPD) medium to OD600 cell densities of 2–2.2 (Supplementary Figure S4B).
Prochlorococcus genes do not complement mutant yeast genes
Cell spotting of a yeast control strain containing only the pmycYACTnNotI vector and spotting of Prochlorococcus yeast clones 1–13, 2–2 and 2–19 on plates lacking histidine, and uracil, leucine or tryptophan (standard marker mutations in the host strain W303-1A) yielded no growth. Growth was normal on plates lacking only histidine or histidine and lysine, thus the bacterial genes for uracil, leucine and tryptophan synthesis do not complement the yeast host genotype (Supplementary Figure S6).
DISCUSSION
To our knowledge this is the first report of the cloning of a gram-negative bacterial genome in S. cerevisiae and the second report of cloning a complete circular genome that uses the standard genetic code (18). DNA sequence analysis showed that the complete MED4 genome was present in the yeast clones. We observed a few mutational differences (primarily single base substitutions) between two sequences of the native Prochlorococcus MED4 genome and a yeast clone of that genome. However, because MED4 is maintained by serial passage without clonal isolation, there is no reason to believe that any of these differences arose by mutation during propagation in yeast.
There are two major reasons that the Prochlorococcus genome may be sustainable in yeast. First, it has a high incidence of ACS sites due to its high AT content, and these may be active as yeast origins of replication. Second, it is likely that most of the Prochlorococcus genes are not accurately expressed in yeast due to differences between the mechanisms of prokaryotic and eukaryotic transcription and translation. Prokaryotic promoters generally contain defined −35 and −10 regions, typically within 50 bp of the start of a gene. However, transcription in cyanobacteria may be more complicated due to additional RNA polymerase subunits γ and δ. Analysis of transcription start sites in Prochlorococcus MED4 has shown similar upstream elements to those of yeast (e.g. TATAAT or TATTAT) (28–30). Thus, transcription initiation of the prokaryotic genome in yeast might be possible. However, other studies involving vaccine development in yeast have shown that transcripts of foreign AT-rich genes are prematurely polyadenylated at unexpected sites (31). The native 1400 bp fragment C gene, a subunit of the tetanus toxin derived from the bacterium Clostridium tetani (GC content 28.6%), could not be expressed in its entirety from a yeast GAL promoter until a synthetic version was engineered with a GC content of 47%. Judging from previous studies expressing constructs up to 3.5 kb, sequences with a GC content below 40% resulted in truncated mRNA transcripts (32).
Translation of Prochlorococcus transcripts would require a high degree of similarity between the ribosome binding sites (Shine–Dalgarno sequences) and the yeast translation initiator consensus sites. This does not seem to be the case for cyanobacteria and yeast. Shine–Dalgarno (S–D) sequences typically have a consensus sequence GGAGG or a degenerate version of this which is within 5–10 bp upstream of the translation start codon. Cyanobacterial and chloroplast genes show very few ribosome binding sites in their 5′-UTR regions that can be classified as S–D consensus sequences even though the 16S rRNA in these organisms still retains the complementary (CCUCC) to the S–D consensus sequence (33,34). While the translation initiation site in polycistronic prokaryotic mRNAs is usually selected by base pairing of the S–D sequence with ribosomal RNA, initiation in eukaryotes is facilitated by a scanning mechanism whereby the small 40 S ribosomal subunit and additional co-factors (eIF2, eIF3, eIF4C, Met-tRNAi and Guanosine-5′-triphospate (GTP)) bind the 5′-cap structure of the mRNA and then migrate down the untranslated leader sequence scanning for the first AUG codon (35,36). Translation initiation of prokaryotic mRNA in yeast could be misplaced toward downstream AUG codons.
Additionally, there may be translational differences due to differences in codon usage between Prochlorococcus and yeast. Although the initiation step of mRNA translation is considered rate-limiting, codon usage is known to affect elongation rate and can be a limiting factor in product yield. Codon usage tables showed a bias in codon usage between Prochlorococcus MED4 and S. cerevisiae. For example, MED4 uses the glycine GGA codon and the leucine codon TTA twice as much compared to yeast (Supplementary Figure S7).
We did not observe a toxic effect as far as growth rates of these yeast clones are concerned. Deep sequencing analysis confirmed the presence of all Prochlorococcus genes inside the yeast host; however, several bacterial genes did not complement the yeast host genotype. We conclude that there is limited expression of Prochlorococcus genes in yeast and that the Prochlorococcus genome is stably maintained under selective conditions as observed for other bacterial genomes cloned in yeast thus far. We see these results as a proof of concept that bacteria using the standard genetic code which remain difficult to isolate or transform can be cloned and maintained in yeast. With the growing number of whole-genome sequences currently available, expanding the range of bacteria that can be co-transformed into yeast is a valuable stepping stone to the study of genetically intractable species.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–11, Supplementary Figures 1–7, Supplementary Chart 1 and Supplementary References [37,38].
FUNDING
Synthetic Genomics Inc. (SGI); all authors were supported by SGI (in part); National Science and Engineering Research Council of Canada (NSERC, Postdoctoral Fellowship) and SGI [to B.J.K.]. Funding for open access charge: JCVI.
Conflict of interest statement. J.C.V. is a Chief Executive Officer and Co-Chief Scientific Officer of Synthetic Genomics Inc. (SGI). H.O.S. is a Co-Chief Scientific Officer and a member of the Board of Directors of SGI. C.A.H. is a Chairman of the SGI Scientific Advisory Board. D.G.G. is a Principal Scientist at SGI. All four of these authors and the J. Craig Venter Institute hold SGI stock.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Yo Suzuki and Philip D. Weyman for critically reading the article and Sanjay Vashee, Mikkel Algire, Radha Krishnakumar, Vladimir Noskov and Chuck Merryman for helpful discussions and suggestions.
REFERENCES
- 1.DiTullio GR, Geesey ME, Jones DR, Daly KL, Campbell L, Smith W., Jr Phytoplankton assemblage structure and primary productivity along 170 W in the South Pacific Ocean. Mar. Ecol. Prog. Ser. 2003;255:55–80. [Google Scholar]
- 2.Chisholm SW, Olson RJ, Zettler ER, Goericke R, Waterbury JB, Welschmeyer NA. A novel free-living prochlorophyte abundant in the oceanic euphotic zone. Nature. 1988;334:340–343. [Google Scholar]
- 3.Partensky F, Hess WR, Vaulot D. Prochlorococcus, a marine photosynthetic prokaryote of global significance. Microbiol. Mol. Biol. Rev. 1999;63:106–127. doi: 10.1128/mmbr.63.1.106-127.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rocap G, Larimer FW, Lamerdin J, Malfatti S, Chain P, Ahlgren NA, Arellano A, Coleman M, Hauser L, Hess WR, et al. Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature. 2003;424:1042–1047. doi: 10.1038/nature01947. [DOI] [PubMed] [Google Scholar]
- 5.Williamson SJ, Rusch DB, Yooseph S, Halpern AL, Heidelberg KB, Glass JI, Andrews-Pfannkoch C, Fadrosh D, Miller CS, Sutton G, et al. The Sorcerer II Global Ocean Sampling Expedition: metagenomic characterization of viruses within aquatic microbial samples. PLoS one. 2008;3:e1456. doi: 10.1371/journal.pone.0001456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rusch DB, Martiny AC, Dupont CL, Halpern AL, Venter JC. Characterization of Prochlorococcus clades from iron-depleted oceanic regions. Proc. Natl Acad. Sci. USA. 2010;107:16184–16189. doi: 10.1073/pnas.1009513107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tolonen AC, Liszt GB, Hess WR. Genetic manipulation of Prochlorococcus strain MIT9313: green fluorescent protein expression from an RSF1010 plasmid and Tn5 transposition. Appl. Environ. Microbiol. 2006;72:7607–7613. doi: 10.1128/AEM.02034-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coleman ML, Sullivan MB, Martiny AC, Steglich C, Barry K, Delong EF, Chisholm SW. Genomic islands and the ecology and evolution of Prochlorococcus. Science. 2006;311:1768–1770. doi: 10.1126/science.1122050. [DOI] [PubMed] [Google Scholar]
- 9.Kettler GC, Martiny AC, Huang K, Zucker J, Coleman ML, Rodrigue S, Chen F, Lapidus A, Ferriera S, Johnson J, et al. Patterns and implications of gene gain and loss in the evolution of Prochlorococcus. PLoS Genet. 2007;3:e231. doi: 10.1371/journal.pgen.0030231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lartigue C, Glass JI, Alperovich N, Pieper R, Parmar PP, Hutchison CA, III, Smith HO, Venter JC. Genome transplantation in bacteria: changing one species to another. Science. 2007;317:632–638. doi: 10.1126/science.1144622. [DOI] [PubMed] [Google Scholar]
- 11.Lartigue C, Vashee S, Algire MA, Chuang R-Y, Benders GA, Ma L, Noskov VN, Denisova EA, Gibson DG, Assad-Garcia N, et al. Creating bacterial strains from genomes that have been cloned and engineered in yeast. Science. 2009;325:1693–1696. doi: 10.1126/science.1173759. [DOI] [PubMed] [Google Scholar]
- 12.Gibson DG, Benders GA, Andrews-Pfannkoch C, Denisova EA, Baden-Tillson H, Zaveri J, Stockwell TB, Brownley A, Thomas DW, Algire MA, et al. Complete chemical synthesis, assembly, and cloning of a Mycoplasma genitalium genome. Science. 2008;319:1215–1220. doi: 10.1126/science.1151721. [DOI] [PubMed] [Google Scholar]
- 13.Gibson DG, Glass JI, Lartigue C, Noskov VN, Chuang R-Y, Algire MA, Benders GA, Montague MG, Ma L, Moodie MM, et al. Creation of a bacterial cell controlled by a chemically synthesized genome. Science. 2010;329:52–56. doi: 10.1126/science.1190719. [DOI] [PubMed] [Google Scholar]
- 14.Benders G, Noskov VN, Denisova EA, Lartigue C, Gibson DG, Assad-Garcia N, Chuang R-Y, Carrera W, Moodie M, Algire MA, et al. Cloning whole bacterial genomes in yeast. Nucleic Acids Res. 2010;38:2558–2569. doi: 10.1093/nar/gkq119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Noskov VN, Segall-Shapiro TH, Chuang R-Y. Tandem repeat coupled with endonuclease cleavage (TREC): a seamless modification tool for genome engineering in yeast. Nucleic Acids Res. 2010;38:2570–2576. doi: 10.1093/nar/gkq099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kouprina N, Larionov V. Selective isolation of genomic loci from complex genomes by transformation-associated recombination cloning in the yeast Saccharomyces cerevisiae. Nat. Protoc. 2008;3:371–377. doi: 10.1038/nprot.2008.5. [DOI] [PubMed] [Google Scholar]
- 17.Noskov VN, Chuang R-Y, Gibson DG, Leem S-H, Larionov V, Kouprina N. Isolation of circular yeast artificial chromosomes for synthetic biology and functional genomics studies. Nat. Protoc. 2010;6:89–96. doi: 10.1038/nprot.2010.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karas BJ, Tagwerker C, Yonemoto IT, Hutchison CA, III, Smith HO. Cloning the Acholeplasma laidlawii PG-8A genome in Saccharomyces cerevisiae as a yeast centromeric plasmid. ACS Synth. Biol. 2012;1:22–28. doi: 10.1021/sb200013j. [DOI] [PubMed] [Google Scholar]
- 19.Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL. Versatile and open software for comparing large genomes. Genome Biol. 2004;5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Palzkill TG, Newlon CS. A yeast replication origin consists of multiple copies of a small conserved sequence. Cell. 1988;53:441–450. doi: 10.1016/0092-8674(88)90164-x. [DOI] [PubMed] [Google Scholar]
- 21.Celniker SE, Sweder K, Srienc F, Bailey JE, Campbell JL. Deletion mutations affecting autonomously replicating sequence ARSI of Saccharomyces cerevisiae. Construction. 1984;4:2455–2466. doi: 10.1128/mcb.4.11.2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nieduszynski CA, Knox Y, Donaldson AD. Genome-wide identification of replication origins in yeast by comparative genomics. Genes Dev. 2006;20:1874–1879. doi: 10.1101/gad.385306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Theis JF, Newlon CS. The ARS309 chromosomal replicator of Saccharomyces cerevisiae depends on an exceptional ARS consensus sequence. Proc. Natl Acad. Sci. USA. 1997;94:10786–10791. doi: 10.1073/pnas.94.20.10786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wyrick JJ, Aparicio JG, Chen T, Barnett JD, Jennings EG, Young RA, Bell SP, Aparicio OM. Genome-wide distribution of ORC and MCM proteins in S. cerevisiae: high-resolution mapping of replication origins. Science. 2001;294:2357–2360. doi: 10.1126/science.1066101. [DOI] [PubMed] [Google Scholar]
- 25.Xu W, Aparicio JG, Aparicio OM, Tavaré S. Genome-wide mapping of ORC and Mcm2p binding sites on tiling arrays and identification of essential ARS consensus sequences in S. cerevisiae. BMC Genomics. 2006;7:276. doi: 10.1186/1471-2164-7-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Noskov VN, Karas BJ, Young L, Chuang R-Y, Gibson DG, Lin Y-C, Stam J, Yonemoto IT, Suzuki Y, Andrews-Pfannkoch C, et al. Assembly of large, high G+C bacterial DNA fragments in yeast. ACS Synth. Biol. 2012;1:267–273. doi: 10.1021/sb3000194. [DOI] [PubMed] [Google Scholar]
- 27.Osburne MS, Holmbeck BM, Frias-Lopez J, Steen R, Huang K, Kelly L, Coe A, Waraska K, Gagne A, Chisholm SW. UV hyper-resistance in Prochlorococcus MED4 results from a single base pair deletion just upstream of an operon encoding nudix hydrolase and photolyase. Environ. Microbiol. 2010;12:1978–1988. doi: 10.1111/j.1462-2920.2010.02203.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vogel J, Axmann I, Herzel H, Hess W. Experimental and computational analysis of transcriptional start sites in the cyanobacterium Prochlorococcus MED4. Nucleic Acids Res. 2003;31:2890–2899. doi: 10.1093/nar/gkg398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sweetser D, Nonet M, Young RA. Prokaryotic and eukaryotic RNA polymerases have homologous core subunits. Proc. Natl Acad. Sci. USA. 1987;84:1192–1196. doi: 10.1073/pnas.84.5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singer V, Wobbe C, Struhl K. A wide variety of DNA sequences can functionally replace a yeast TATA element for transcriptional activation. Genes Dev. 1990;4:636–645. doi: 10.1101/gad.4.4.636. [DOI] [PubMed] [Google Scholar]
- 31.Romanos M, Scorer C, Clare J. Foreign gene expression in yeast: a review. Yeast. 1992;8:423–488. doi: 10.1002/yea.320080602. [DOI] [PubMed] [Google Scholar]
- 32.Romanos M, Makoff A. Expression of tetanus toxin fragment C in yeast: gene synthesis is required to eliminate fortuitous polyadenylation sites in AT-rich DNA. Nucleic Acids Res. 1991;19:1461–1467. doi: 10.1093/nar/19.7.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo Z, Sherman F. 3’-end-forming signals of yeast mRNA. Mol. Cell. Biol. 1995;15:5983–5990. doi: 10.1128/mcb.15.11.5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weinberg Z, Barrick JE, Yao Z, Roth A, Kim JN, Gore J, Wang JX, Lee ER, Block K, Sudarsan N, et al. The CMfinder comparative genomics pipeline discovers 22 novel putative RNAs in bacteria. Supplementary Data: diagrams of all motifs. Nucleic Acids Res. 2007;35:4809–4819. doi: 10.1093/nar/gkm487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karlin S, Mrazek J. Predicted highly expressed genes of diverse prokaryotic genomes. J. Bacteriol. 2000;182:5238–5250. doi: 10.1128/jb.182.18.5238-5250.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kozak M. Initiation of translation in prokaryotes and eukaryotes. Gene. 1999;234:187–208. doi: 10.1016/s0378-1119(99)00210-3. [DOI] [PubMed] [Google Scholar]
- 37.Gibson DG, Benders GA, Axelrod KC, Zaveri J, Algire MA, Moodie M, Montague MG, Venter JC, Smith HO, Hutchison CAIII. One-step assembly in yeast of 25 overlapping DNA fragments to form a complete synthetic Mycoplasma genitalium genome. Proc. Natl Acad. Sci. USA. 2008;105:20404–20409. doi: 10.1073/pnas.0811011106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fuhrmann M, Hausherr A, Ferbitz L, Schödl T, Heitzer M, Hegemann P. Monitoring dynamic expression of nuclear genes in Chlamydomonas reinhardtii by using a synthetic luciferase reporter gene. Plant Mol. Biol. 2004;55:869–881. doi: 10.1007/s11103-004-2150-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.