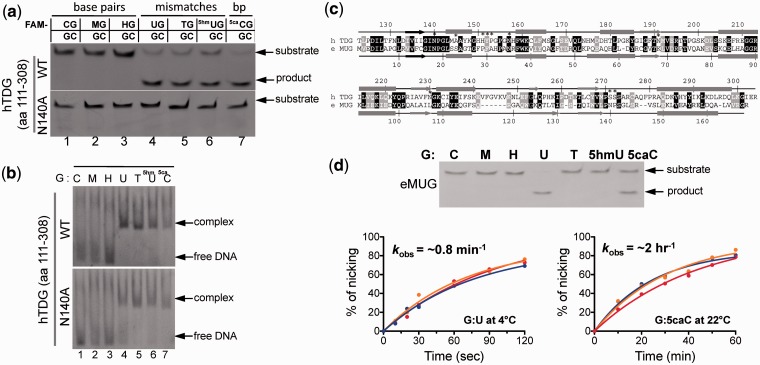
Figure 2.
Base excision and binding activities of TDG catalytic domain in the context of a double-stranded CpG dinucleotide. (a) Double-stranded 32-bp oligonucleotides bearing a single CpG dinucleotide were incubated with equal amount of the glycosylase domain of TDG or its noncatalytic mutant N140A at 37°C for 30 min. The oligonucleotide was labeled with FAM on the top strand, and the modification status was indicated (M = 5mC and H = 5hmC). The products of the reactions were separated on a denaturing polyacrylamide gel, and the FAM-labeled strand was excited by UV and photographed. (b) DNA binding assays were performed by incubating 0.5 µM FAM-labeled oligonucleotides with 1 µM of TDG at 37°C for 15 min. (c) Pairwise sequence alignment of human TDG domain (top line) and E. coli MUG (bottom line). Secondary structural elements are shown above or below the aligned sequences. White-on-black residues are invariant between the two sequences examined, while gray-highlighted positions are conserved (R and K, E and D, Q and N, T and S, F, Y and W, V, I, L and M, and G and P). Positions highlighted by * are active site residues responsible for catalysis (Asn140) and/or proposed for substrate base recognition (only two of them, Asn140 and Asn157, are invariant between human TDG and E. coli MUG). (d) eMUG is active on G:U and G:5caC substrates (top panel). Reactions were performed at room temperature (approximately 22°C) for 30 min with [EeMUG] = [SDNA] = 5 µM. The kinetic activities of eMUG on G:U substrates at 4°C (bottom left panel) and G:5caC at room temperature (approximately 22°C) (bottom right panel) were measured under single turnover condition ([EeMUG] = 2.5 µM and [SDNA] = 0.25 µM) at three different pH values (5.5 in red, 7.5 in orange and 8.0 in blue curves).