Figure 4.
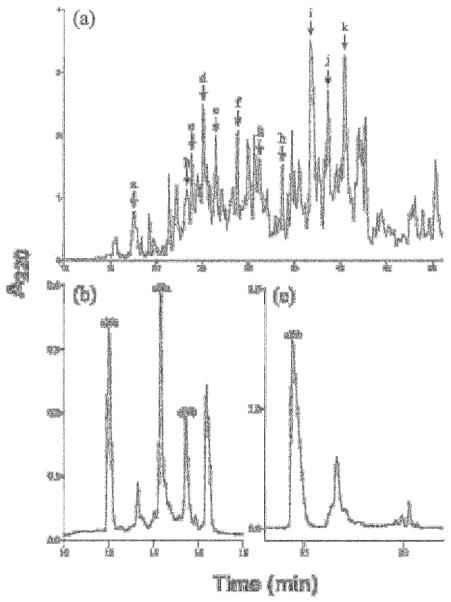
Identification of conopeptides from venom fractions of Conus californicus. (a) Fractionation of C. californicus venom extract on a Vydac C18 analytical column with a linear gradient of 1.0% B90/min at a flow rate of 1.0 mL/min. Lettered arrows represent HPLC fractions used to identify primary amino acid sequences depicted in Table 1. (b) Subfractionation of peak c from panel (a), after reduction with 15 mM DDT and alkylation with 4-vinylpyridine using an elution gradient of 15-40% B90 over 30 minutes; three peptides, cl 6a, cl 9a and cl 9b were sequenced from this fractionation (c) Fractionation of peak i from panel (a), leading to the characterization of cl 6b the most abundant peptide within the venom of Conus californicus, using a Vydac C18 analytical column eluded with a gradient of 15-60% buffer B90 over 60 min. The sequences of the four peptides marked in panels (b) and (c) were deduced from an automated Edman degredation and are presented in Table 1.
