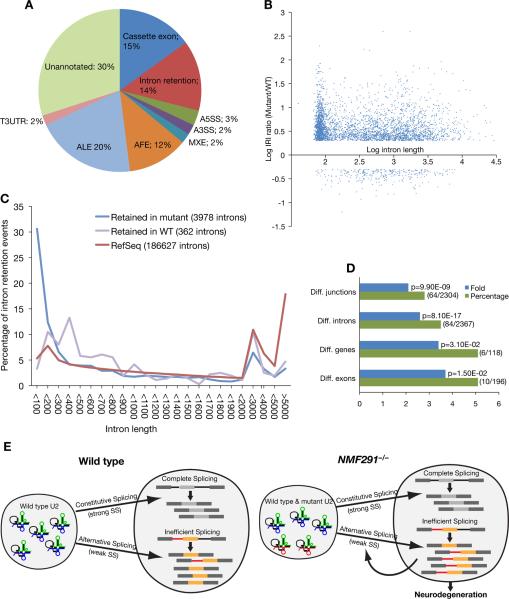
Figure 7. Global Splicing Abnormalities in the NMF291−/− Cerebellum.
(A) Types of alternative splicing events identified by exon array analysis as differentially spliced between the wild type and the NMF291−/− cerebellum. A3SS, alternative 3' splice site; A5SS; alternative 5' splice site; AFE, alternative first exon; ALE, alternative last exon; MXE, mutally exclusive exon; T3UTR, truncated 3'UTR.
(B) The log of the mutant/WT Relative Intron retention Index (IRI) generated by RNA-Seq analysis plotted against the log of the intron length (bp).
(C) Significantly retained introns in the cerebellum of NMF291−/− or wild type mice, or RefSeq introns were grouped by length and plotted as a percentage of the total introns.
(D) Gene Ontology (GO) analysis of genes annotated to the pre-mRNA splicing GO term were significantly enriched in the differentially spliced exons (Diff. junctions) and introns (Diff. introns) identified by RNA-Seq, and differentially spliced exons (Diff. exons) and differentially expressed genes (Diff. genes) identified by exon-array. Percentage, the number of genes (the numerator in parentheses) annotated to the GO term divided by the total number of inputted genes (the denominator in parentheses). Fold enrichment, the magnitude of enrichment of input genes relative to all mouse genes annotated to pre-mRNA splicing. p-value, the significance of gene-term enrichment with a modified Fisher's exact test.
(E) The working model for our findings. The splicing status and biological consequences of expression of wild type (blue) and NMF291 mutant (red) U2 snRNAs in wild type and NMF291−/− neurons are shown. Our in vivo and in vitro data suggest that expression of mutant U2 snRNA disrupts splicing at many suboptimal or weaker splice sites, including some of those used in alternative splicing. These changes in alternative splicing may be further amplified by altered splicing of RNA processing factors caused directly by mutant U2 snRNAs or by autoregulation. See also Figure S6 and Table S1A–H.