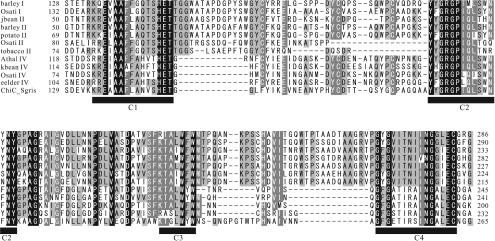
FIG. 2.
Alignment of the conserved regions of family 19 chitinases used to design PCR primers. Residues conserved in all sequences are indicated by white type on a black background, while residues conserved in >80 and >60% of the proteins examined are indicated by white type on a dark gray background and by black type on a light gray background, respectively. Conserved regions of family 19 chitinases are indicated by the solid bars (C1, C2, C3, and C4). Conserved regions 1 and 2 were used to design PCR primers. barley I, barley class I chitinase (amino acid sequence accession no. Q42839); Osati I, O. sativa class I chitinase (Q42992); jbean II, jack bean class II chitinase (O81934); barley II, barley class II chitinase (P11955); potato II, potato class II chitinase (Q43184); Osati II, O. sativa class II chitinase (O80423); tobacco II, tobacco class II chitinase (P17514); Athal IV, Arabidopsis thaliana class IV chitinase (O23248); kbean IV, kidney bean class IV chitinase (P27054); Osati IV, O. sativa class IV chitinase (O04138); eelder IV, European elder class IV chitinase (Q43150); ChiC_Sgris, S. griseus HUT6037 ChiC (O50152).