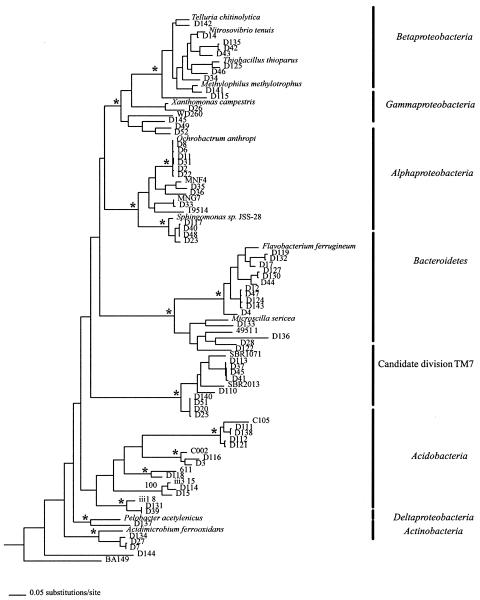
FIG. 2.
A 50% majority rule consensus tree of 16S rRNA genes derived from Bayesian phylogenetics. An asterisk indicates a node with a Bayesian posterior probability of >0.95, maximum parsimony bootstrap support of >80, and neighbor-joining bootstrap support of >80. The sequences from this work are the D series. The tree is rooted with Methanosarcina acetivorans (M59137) and Natronobacterium chahannaoensis (AJ004806). Branch lengths are drawn proportional to the amount of evolution based on uncorrected genetic distances. Accession numbers are as follows: Telluria chitinolytica, X65590; Nitrosovibrio tenuis, M96405; Thiobacillus thioparus, M79426; Methylophilus methylotrophus, L15475; X. campestris ATTC, 339113; WD260, AJ292673; O. anthropi, D63837; MNF4, AF292996; MNG7, AF292997; 19514, AF097791; Sphingomonas sp. strain JSS-28, AF031240; Flavobacterium ferrugineum, M28237; Microscilla sericea, M58794; 49511, AF097805; SBR1071, AF268996; SBR2013, AF269000; C105, AF013530; C002, AF013515; 611, Y11629; ii3_15, Z95725; iii1_8, Z95729; Pelobacter acetylenicus, X70955; Acidimicrobium ferrooxidans, U75647; BA149, AF323777.