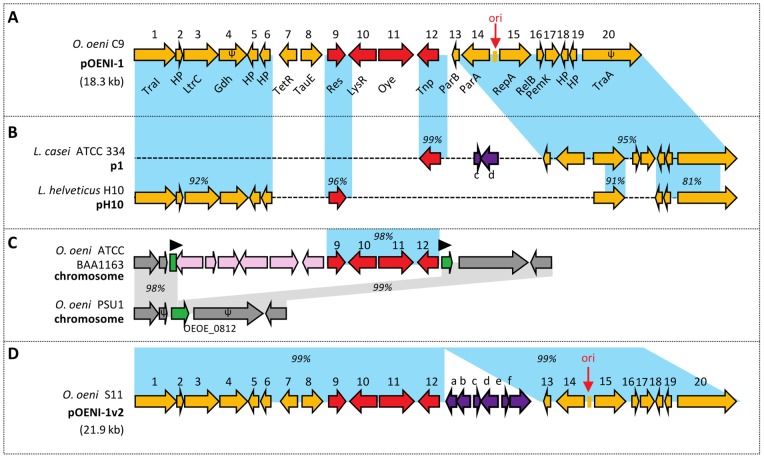
Figure 1. Genetic organization of pOENI-1 and comparison with related sequences.
A. Genetic organization of plasmid pOENI-1. ORFs are represented by numbered arrows and identified by corresponding protein tags (see also Table 3). B. Sequence comparison of pOENI-1 and related plasmids p1 (CP000424) and pH10 (CP002430). ORFs “c, d” (purple arrows) share 99% similarity with ORFs of pOENI-1v2. C. Portions of chromosomes in O. oeni ATCC BAA 1163 and O. oeni PSU1. The gene OEOE_0812 in O. oeni PSU1 (green arrow) is disrupted in O. oeni ATCC BAA 1163 by an 10 genes insert comprising four genes conserved in pOENI-1 (red arrows) and six genes unrelated to pOENI-1 (pink arrows). The insert is bordered by an 8-bp repeated sequence (dark triangles). D. Genetic organization of pOENI-1v2. ORFs numbered from 1 to 20 share more than 99% nucleotide sequence similarity with corresponding ORFs in pOENI-1. ORFs shaded in purple are not detected in pOENI-1 and code for transposases (a, e, f,), hypothetical proteins (b, c) and a recombinase (d). Pseudogenes are symbolized by arrowheads containing the symbol ψ. Regions of sequence similarity are indicated in percentages and shaded in blue. ori: putative origin of replication.