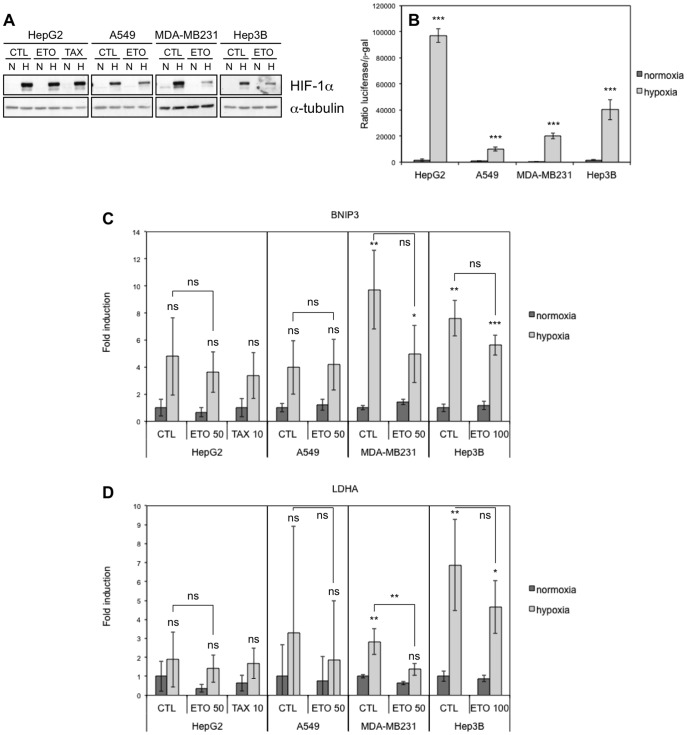
Figure 3. Effect of hypoxia, etoposide and paclitaxel on HIF-1alpha abundance and HIF-1 activity.
HepG2, A549, MDA-MB231 and Hep3B cells were incubated 16 hours under normoxia (N, 21% O2) or hypoxia (H, 1% O2) (A-D) in the presence or not (CTL) of etoposide (ETO, 100 µM in Hep3B cells and 50 µM in the other cell types) or paclitaxel (TAX, 10 µM) in HepG2 cells (A, C, D). (A) HIF-1alpha was detected in total cell extracts by western blotting using a specific antibody. Alpha-tubulin was used as loading control. One experiment representative out of three. Uncropped western blots are presented in Figure S1. (B) Before incubation, cells were co-transfected with the pGL3-(PGK-HRE6)-tk-luc reporter plasmid encoding the firefly luciferase and the pCMVß normalisation plasmid. Results are expressed as mean of the ratio between firefly luciferase activity and ß-galactosidase activity ±1 SD (n = 3). Statistical analysis was carried out with ANOVA 1. ***: P<0.001. Symbols are a comparison between the normoxic and hypoxic conditions of the same cell type. (C, D) After incubation, total RNA was extracted, submitted to reverse transcription and then to amplification in the presence of SYBR Green and specific primers for BNIP3 (c) or LDHA (D). RPL13A was used as housekeeping gene for data normalization. Data are given in fold-induction as the mean ±1 SD (n = 3). Statistical analysis was carried out with ANOVA 1. ns: non-significant; *: P<0.05; **: P<0.01; ***: P<0.001. Symbols above the hypoxic columns are a comparison with the corresponding normoxic condition (with the same drug).