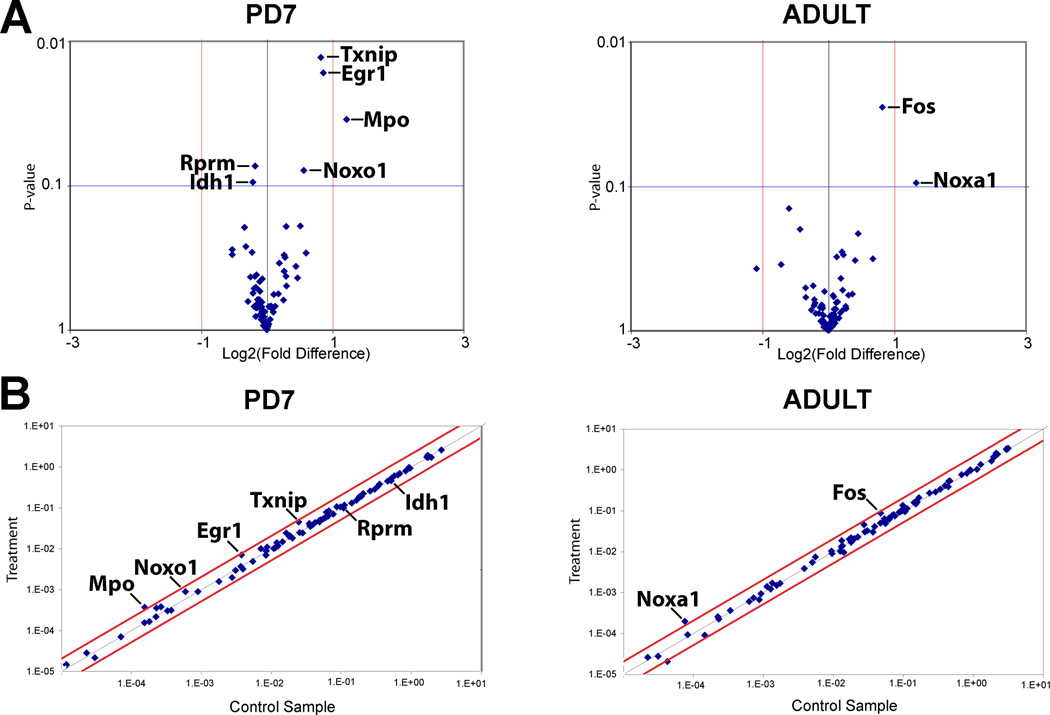
Figure 3. Gene Expression Analysis at the Ventral Periaqueductal Gray.
Total of 84 genes were assayed using Rat Nitric Oxide Signaling Pathway PCR Array (PARN-062; SABiosciences™) in in the ventral periaqueductal gray at the level of the inferior colliculus (see also Fig. 1B). Gene expression was compared between treatment (chronic morphine) and control (chronic normal saline administration) at two different ages of the rat: postnatal day (PD)7 and adult. (A) Volcano plots illustrate relationship between the gene expression fold change and p-value for all the morphine tolerant and control pairs. Those with p < 0.1 for treatment effect are indicated. (B) Scatter plots for all of the genes assayed illustrating the relative abundance in control and chronic morphine treated conditions. X and Y axis in Panel B correspond to 2^-ΔCt [Ct(gene of interest)-Avg Ct (housekeeping gene, HKG]. Abbreviations of genes: Egr1, Early growth response 1; Fos, FBJ osteosarcoma oncogene; Idh1, isocitrate dehydrogenase 1 (NADP+), soluble; MPO, myeloperoxidase; Noxa1, NADPH oxidase activator 1; Noxo1, NADPH oxidase organizer 1; Txnip, thioredoxin interacting protein.