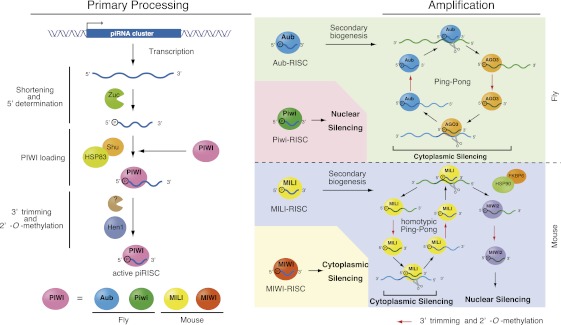
Figure 1.
piRNA biogenesis in Drosophila and mice consists of the primary piRNA processing pathway and the amplification loop. The left side shows a model of the primary processing pathway in flies. The primary transcripts of piRNA clusters are shortened into piRNA-like small RNAs. The factors necessary for this process largely remain to be determined. Recent studies have suggested that Zucchini might be an endonuclease forming the 5′ ends of piRNAs. The mouse Zucchini homolog (MitoPLD) might also function as a nuclease to produce primary piRNAs. piRNA-like small RNAs are loaded onto PIWI proteins (shown in pink) and then trimmed from the 3′ end to the size of mature piRNAs by an unknown nuclease. Hsp83/Shu may play a role in the Piwi loading step. piRNAs are 2′-O-methylated by HEN1/Pimet. In flies, the PIWI proteins that associate with primary piRNAs are Piwi (green) and Aub (blue). Piwi associated with piRNAs is translocated to the nucleus and thus likely does not contribute to the amplification loop. Aub associated with piRNAs now triggers the Ping-Pong cycle by cleaving transposon transcripts. Ago3 (orange) loaded with secondary piRNAs in turn produces piRNAs that associate with Aub. Transposon transcripts are cleaved through this amplification cycle, resulting in cytoplasmic transposon silencing. In mice, primary piRNAs are loaded onto MILI (yellow) and MIWI (red). MIWI associated with pachytene piRNAs functions in cytoplasmic silencing. The targets remain largely unknown. MILI associated with primary piRNAs contributes to the Ping-Pong cycle to produce piRNAs that associate with MIWI2 (purple), upon which MIWI2 is localized to the nucleus to accomplish nuclear silencing. The contribution of MIWI2 to the Ping-Pong cycle may be negligible. MILI might operate a homotypic Ping-Pong cycle as indicated. HSP90/FKBP6 plays a role in producing secondary piRNAs that associate with MIWI2. HEN1/Pimet 2′-O-methylates secondary piRNAs, the products of the Ping-Pong cycle.