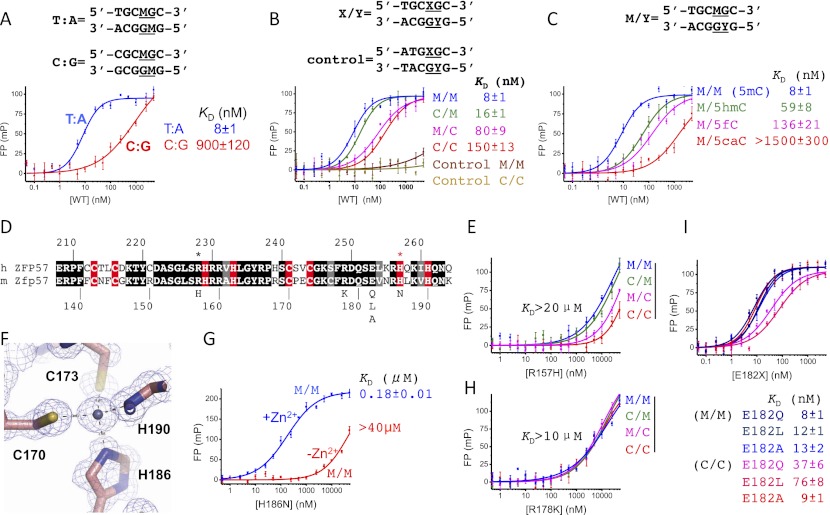
Figure 3.
The effects of DNA sequence, 5mC oxidation, and Zfp57 mutants on DNA binding. (A) The first T:A base pair of the TGCCGC element is important for recognition by Zfp57. (B) Binding affinities between ZF2–3 and TGCCGC with four different modification states (M/M, C/M, M/C, and C/C, where M is 5mC and C is Cyt). A 5′ half-mutated sequence (ATGCGC) was used for negative control. (C) The effect of 5mC oxidation ([5hmC] 5-hydroxymethylation; [5fC] 5-formylation; [5caC] 5-carboxylation) on DNA binding. (D) The sequence alignment between human ZFP57 and mouse Zfp57, with white-on-black residues being invariant among the corresponding regions of sequences examined, gray-highlighted positions conserved, and white-on-red being zinc ligands. (E) The effect of the R157H mutant on DNA binding. Although there is significantly reduced affinity, fully methylated DNA (M/M) is still preferred over hemimethylated DNA (C/M and M/C) and unmodified DNA (C/C) by this mutant. (F) His186 is one of the zinc ligands. (G) The effect of Zn2+ (1 mM ZnCl2 added in the reaction buffer) on DNA binding by the H186N mutant. (H) R178K loses the discrimination of methylation status. (I) E182 is dispensable for binding methylated DNA.