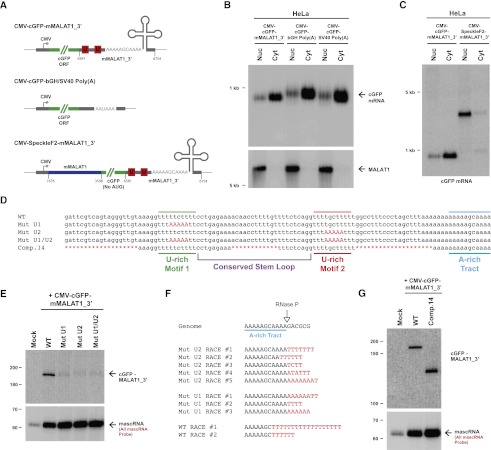
Figure 2.
The U-rich motifs inhibit uridylation and degradation of the 3′ end of MALAT1. (A) Schematics of cGFP expression plasmids used in this study. (Middle) To generate a cGFP transcript ending in a canonical poly(A) tail, the mMALAT1_3′ region was replaced with either the bovine growth hormone (bGH) or the SV40 polyadenylation signal. (Bottom) To generate a nuclear-retained cGFP transcript, nucleotides 1676–3598 of mMALAT1 was placed upstream of cGFP. (B) Transfected HeLa cells were fractionated to isolate nuclear and cytoplasmic total RNA, which was then subjected to Northern blot analysis with a probe to the cGFP ORF. A probe to endogenous MALAT1 was used as a control for fractionation efficiency. (C) The SpeckleF2-MALAT1_3′ transcript was efficiently retained in the nucleus. (D) Mutations or deletions (denoted in red) were introduced into the mMALAT1_3′ region of the CMV-cGFP-mMALAT1_3′ expression plasmid. (E) The wild-type (WT) or mutant plasmids were transfected into HeLa cells, and Northern blots were performed. RNase H treatment was performed prior to the Northern blot that detected cGFP-MALAT1_3′ RNA. (F) A ligation-mediated 3′ RACE approach was used to examine the 3′ ends of cGFP-MALAT1_3′ transcripts undergoing degradation. Nucleotides added post-transcriptionally are in red. (G) RNase H treatment followed by Northern blotting was used to show that the cGFP-MALAT1_3′ Comp.14 transcript is stable. As 51 nt were deleted to generate the Comp.14 transcript, a band of only 139 nt is expected.