Abstract
Regulation of dendritically localized mRNAs offers an important means by which neurons can sculpt precise signals at synapses. Arc is one such dendritically localized mRNA, and it has been shown to contain two exon-junction complexes (EJC) within its 3’UTR. The EJC has been postulated to regulate cytoplasmic Arc mRNA availability through translation dependent decay and thus contribute to synaptic plasticity. Core proteins of the EJC include eIF4A3, an RNA helicase, and Magoh, which stabilizes the interaction of eIF4A3 with target mRNAs. Arc mRNA expression is activity-regulated in numerous brain regions, including the dorsal striatum and hippocampus. Therefore in this study, the in vivo expression of these core EJC components was investigated in adult Sprague-Dawley rats to determine whether there are also behaviorally-regulated changes in their expression. In the present work, there was no change in the expression of Magoh mRNA following spatial exploration, a paradigm previously reported to robustly and reliably upregulate Arc mRNA expression. Interestingly, however, there were increases in eIF4A3 mRNA levels in dorsal striatum and hippocampus following spatial exploration, similar to previous reports for Arc mRNA. Furthermore, there were activity-dependent changes in eIF4A3 protein distribution and expression within striatum following spatial exploration. Importantly, eIF4A3 protein colocalized with Arc mRNA in vivo. Like Arc mRNA expression, eIF4A3 mRNA expression in dorsomedial striatum, but not dorsolateral striatum or hippocampus, significantly correlated with behavioral performance on a striatally-mediated, response-reversal learning task. This study provides direct evidence that a core EJC component, eIF4A3, shows activity-dependent changes in both mRNA and protein expression in the adult mammalian brain. These findings thus further implicate eIF4A3 as a key mediator of Arc mRNA availability underlying learning and memory processes in vivo.
Keywords: exon-junction complex, eIF4A3, striatum, T-maze, rat, Arc/Arg3.1
1
Many dendritically localized mRNAs, including the immediate-early gene Arc/Arg3.1 (activity-regulated, cytoskeleton-associated protein), are subjected to numerous types of intraneuronal processing, including post-transcriptional regulation and localized protein synthesis (Ule and Darnell, 2006, Panja et al., 2009). Such post-transcriptional processing plays a key role in mediating normal, synapse-specific plasticity (Klann and Dever, 2004, Sutton and Schuman, 2006). The exon-junction complex (EJC) is critical to global mRNA function (Tange et al., 2004), with a role mediating neuronal Arc mRNA expression recently coming to light (Giorgi et al., 2007). One postulated mechanism regulating Arc mRNA availability is translation dependent decay (TDD) through the RNA surveillance process of nonsense-mediated mRNA decay (NMD) (Giorgi et al., 2007, Soule et al., 2012), wherein mRNAs can be degraded following the first round of translation if EJCs remain bound to the transcript (Maquat, 2004). Arc mRNA is a natural target for TDD due to the two EJCs within the 3’UTR, theoretically leading to tight control of Arc mRNA availability and protein synthesis (Giorgi et al., 2007). Thus, the EJC and associated mRNA decay processes could potentially contribute to Arc mRNA availability and thus synapse-specific signaling and plasticity.
The minimally stable core of the EJC consists of eIF4A3, Magoh/Y14 and MLN51 (Ballut et al., 2005, Tange et al., 2005); absence of eIF4A3 inhibits EJC deposition (Shibuya et al., 2004). As eIF4A3 is the keystone mRNA binding protein of the EJC, it may act as a brake on cytoplasmic Arc mRNA availability (Giorgi et al., 2007) and thereby potentially play a key role in neuroplasticity. Dynamic regulation of eIF4A3 may thus offer an intriguing means to modulate synaptic plasticity processes in the adult mammalian brain due to its core role in EJC formation (Shibuya et al., 2004, Tange et al., 2005) and known association with mRNAs critical to neuronal function (Giorgi et al., 2007). However, whether eIF4A3 mRNA and protein expression changes in response to neuronal activation in the adult mammalian brain in vivo has yet to be fully explored and was thus the goal of the present work.
In the present study, we demonstrate activity-dependent increases in the expression of eIF4A3 mRNA, but not the expression of another EJC component Magoh, in the adult rodent CNS. We also report brain-region specific relation of eIF4A3 mRNA expression to behavioral indices of new learning, consistent with previous observations for Arc mRNA (Guzowski et al., 1999, Daberkow et al., 2007, 2008). The present findings thus suggest that the expression of eIF4A3 is activity-regulated and that eIF4A3 expression in brain regions being engaged in a particular learning task correlates with that learning. These results thus provide evidence that not only do neurons regulate many effector genes critical for plasticity processes, they also dynamically regulate the regulatory elements contributing to such signaling in the adult mammalian brain in vivo.
2. Experimental Procedures
2.1. Animals
Male Sprague-Dawley rats (Charles River Laboratories, Raleigh, NC; 275-300 g) were singly housed in tub cages in a room controlled for temperature and lighting (12:12 hr). Animal care and experimental procedures conformed to the Guide for the Care and Use of Laboratory Animals (The National Academies Press, 8th Ed.) and were approved by the Institutional Animal Care and Use Committee at the University of Utah.
2.2. Novel Spatial Exploration
Rats were divided into five experimental groups of 4-10 rats each. Rats were handled prior to the experiment to familiarize them to the experimenter and handling. Rats were exposed to a 2-ft × 2-ft plastic tub, novel spatial environment, a paradigm known to robustly and reliably induce Arc mRNA and other immediate-early gene expression, as previously described (Guzowski et al., 1999, Chawla et al., 2005, Vazdarjanova et al., 2006, Daberkow et al., 2007). Rats were removed from their home cage after having been isolated in the cage for 24 hours. Caged control (CC) rats (n=10) were sacrificed immediately upon removal from the home cage. The “5 min” group rats (n=10) were exposed to the novel environment for 5 min and then immediately sacrificed; “30 min” group rats (n=10) were exposed for 5 min to the novel environment and then returned to the home cage for 25 min before sacrifice; “60 min” group rats (n=4) were exposed for 5 min to the novel environment and then returned to the home cage for 55 min before sacrifice. Animals were sacrificed by exposure to CO2, decapitated and brains immediately removed and flash-frozen in 2-methylbutane (Mallinckrodt Baker, Phillipsburg, NJ) chilled on dry ice.
2.3. T-maze, response-reversal learning task
A separate group of rats (n=11) was habituated to the T-maze and experimenter as previously described (Daberkow et al., 2007, 2008, Pastuzyn et al., 2012). After T-maze habituation, the turn bias of each rat was determined, and acquisition training on the T-maze proceeded as previously described (Daberkow et al., 2007, Pastuzyn et al., 2012). During the response-reversal task, rats were rewarded for turning in the opposite direction from acquisition with task completion being when the rat reached criterion (9 / 10 correct consecutive turns). Each trial on this task took approximately 1 min. Five min after reaching criterion, animals were sacrificed and brains collected as described above. As with the novel spatial exploration task, CC rats (n=11) associated with this experiment were immediately sacrificed upon removal from their home cage.
2.4. Tissue preparation
Striatal and hippocampal sections (Bregma: +1.2–1.5 mm and -2.8–3.3 mm, respectively (Paxinos and Watson, 1998)) were cryosectioned at 12-μm (Cambridge Instruments, Bayreuth, Germany) and thaw-mounted onto SuperFrost Plus slides (VWR, Batavia, IL). Slides to be directly compared were processed in parallel. Slides were postfixed as previously described (Ganguly and Keefe, 2001), air-dried and stored at -20°C until histochemical processing.
2.5. Fluorescent immunohistochemistry
Striatal sections to be labeled for eIF4A3 protein were washed 2 × 5 min in PBS/0.1% Triton-X (PBS-T), blocked for 1 hr with PBS/0.1% Triton X/0.1% Bovine Serum Albumin (PBSTB), and then washed 2 × 5 min in PBS-T. Protein was then detected with rabbit anti-eIF4A3 antibody (Abcam, Cambridge, MA) at a concentration of 5 μg/mL overnight at 4°C. Slides were then washed 2 × 5 min in PBS-T followed by incubation with donkey anti-rabbit Alexa Fluor-488 (1:1000; Invitrogen, Carlsbad, CA) in PBS-TB for 2 hr at 24°C. Slides were washed 2 × 5 min in PBS-T and coverslipped with Prolong Gold mounting media with DAPI nuclear counterstain (Invitrogen, Carlsbad, CA).
2.6. Radioactive in situ hybridization
Full-length rat eIF4A3 and mouse Magoh (93% sequence homology to rat) cDNA-containing vectors were purchased for plasmid isolation (eIF4A3 [GenBank: BC105875.1] clone ID 7120292; Magoh [GenBank: BC018176.1] clone ID 3587774; Open Biosystems, Huntsville, AL). The cDNAs were linearized (EcoRI; Roche Applied Science, Indianapolis, IN), and probes transcribed with [S35]-UTP or [P33]-UTP (Perkin Elmer, Waltham, MA) with T7 RNA polymerase (Roche Applied Science, Indianapolis, IN). In situ hybridization was performed as previously described (Ganguly and Keefe, 2001).
2.7 Dual Arc fluorescent in situ hybridization and eIF4A3 immunohistochemistry
Co-expression of Arc mRNA and eIF4A3 protein in dorsal striatum was determined by combined fluorescence in situ hybridization histochemistry (FISH) for Arc mRNA and eIF4A3 protein, as previously described for Arc FISH (Daberkow et al., 2007, 2008) and as described above (2.5) for eIF4A3 protein with minor modifications to the buffers (TNT buffer: 0.1M Tris-HCl, pH 7.5, 0.15M NaCl, 0.05% Tween-20). A full-length ribonucleotide probe complementary to Arc mRNA (Lyford et al., 1995) was synthesized from cDNAs using digoxigenin-UTP (DIG-UTP) with T7 RNA polymerase and DIG-UTP RNA labeling kit (Roche) (Daberkow et al., 2007, 2008). Slides were hybridized with Arc ribonucleotide probe overnight (12–18 h) in a humid chamber at 56 °C. Once removed, slides were vigorously washed at 24°C four times in 2 × SSC buffer (0.15 M NaCl with 0.015 M sodium citrate). Slides were then washed in ribonuclease A (RNase A; 10 μg / mL; Roche Applied Science) in 2 × SSC for 15 min. After incubation with RNase A, slides were washed 5 min in 2 × SSC, then 4 × 20 min in 0.2 × SSC at 24°C. Endogenous peroxidase activity was then quenched with 2% H2O2 for 15 min, slides were washed 2 × 5 min in TNT buffer, and then slides were incubated for 2 hours at RT with an anti-digoxigenin antibody (1:1000) coupled to horseradish peroxidase (HRP; Roche). The Arc probe was detected by cyanine-3 (cy-3) tyramide signal amplification (TSA Plus; Perkin–Elmer). After detection of the DIG-labeled Arc ribonucleotide probe, slides were washed in 2 × 5 min in TNT and eIF4A3 protein detected as above (2.5). The next day, slides were washed 2 × 5 min in TNT, incubated for 2 hours RT with a goat anti-rabbit-HRP antibody (Millipore, Billerica, MA), then washed 2 × 5 min in TNT. The eIF4A3 protein signal was detected by fluorescein TSA (Perkin–Elmer) then washed 2 × 5 min in TNT. Finally, slides were coverslipped with Prolong Gold mounting media with DAPI (Invitrogen).
2.8. Image acquisition and analysis
To determine eIF4A3 protein expression in dorsomedial striatum (+1.2–1.5 mm bregma), 0.6 mm2 fields from two striatal sections per animal were imaged on a Leica DM4000B fluorescent microscope at 40x. Each image was analyzed using ImageJ (NIH) with the inverted LUT threshold set to 15 to remove background. The mean gray value and percent of the total field area with eIF4A3 signal were measured.
Film autoradiograms of sections processed for radioactive in situ hybridization were digitized (ImageJ). For both eIF4A3 and Magoh mRNAs, four sections per animal were imaged from dorsal striatum (rostral (+1.2–1.5 mm bregma) and middle (+0.5 mm bregma)) and dorsal hippocampus (-2.8–3.3 mm bregma). Film autoradiograms were analyzed using ImageJ (Ganguly and Keefe, 2001). The mean gray value of white matter was subtracted from the mean gray value of the regions of interest. Mean gray values for each animal were then normalized to the average signal in the CC group.
Combined FISH/immunohistochemistry images (105.5 μm × 105.5 μm) in dorsomedial striatum (+1.2–1.5 mm bregma; Figure 4A-D) were captured under 2x zoom magnification with an FV1000 confocal laser-scanning microscope (Olympus) with motorized stage (Prior Scientific) using a 60x, 1.45 NA osil-immersion lens (plan APO) and 405-nm Diode, 488-nm Ar, and 543-nm HeNe lasers (Daberkow et al., 2007, 2008). Areas of analysis were z-sectioned in 0.5-μm-thick optical sections.
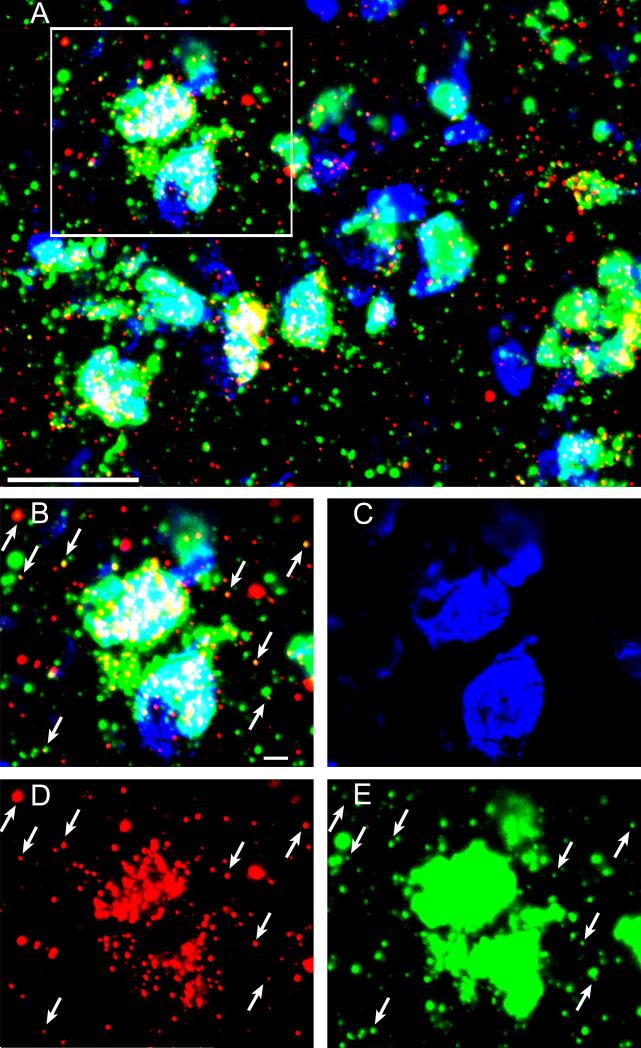
Figure 4. Arc mRNA colocalizes with eIF4A3 protein in vivo following response-reversal learning on a T-maze.
(A) Representative photomicrograph of Arc mRNA in situ hybridization histochemical staining (red) and eIF4A3 protein immunofluorescence (green) in dorsal striatum of a rat sacrificed 5 min after reaching criterion on a striatally-mediated, response-reversal learning task (see Methods). Scale bar = 20 μm. (B-E) Higher magnification images of the region delineated in the box in (A) showing colocalization of eIF4A3 protein expression and Arc mRNA colocalization (B) and the individual channels for the DAPI nuclear counter stain (C), Arc mRNA (D), and eIF4A3 protein (E). Arrows highlight points of colocalization of Arc mRNA and eIF4A3 signal.. Scale bar in B-E = 2 μm.
2.9. Statistical analyses
Expression of mRNAs and protein in rats after spatial exploration were compared using one-way ANOVAs followed by post-hoc Dunnett's tests. Levels of eIF4A3 mRNA expression following response-reversal learning were compared to CC levels using two-tailed t-tests and also correlated with performance.
3. Results
3.1. Spatial Exploration and expression of EJC components eIF4A3 and Magoh
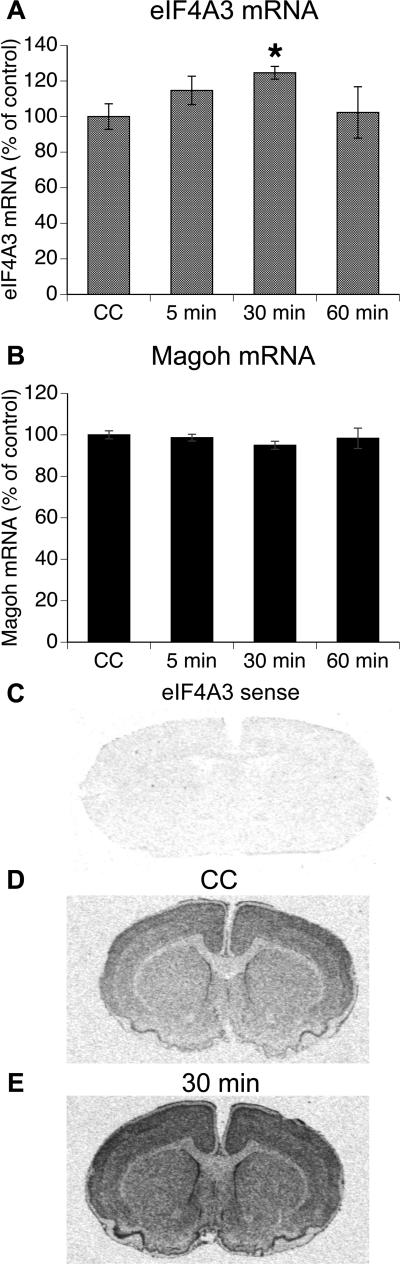
Initially, we characterized the in vivo expression of the EJC factor eIF4A3 in dorsal striatum of rats following spatial exploration of a novel environment (Figure 1). eIF4A3 expression in dorsal striatum increased above that in CC rats following novel environment exploration (Figure 1A; “5 min”=114.7%±8.0 of CC; “30 min”=123.0%±3.9 of CC; “60 min”=102.3%±14.4 of CC; F(3,35)=3.29, p=0.03). Post-hoc analysis revealed a significant increase over CC animals in the “30 min” group (p=0.03; Fig 1D,E). We then asked whether spatial exploration was associated with an increase in Magoh expression, which would suggest global changes in the expression of EJC factors. Although Magoh was also basally expressed, we did not detect any significant increases in Magoh mRNA expression in animals that engaged in the spatial exploration task (“5 min”=98.7%±1.64 of CC; “30 min”=95.9%±1.93 of CC; “60 min”=98.3%±4.91 of CC; F(3,20)=0.73, p>0.05; Figure 1B).
Figure 1. Expression of EJC components in dorsal striatum of rats engaged in spatial exploration for 5 min.
(A) Mean expression (arbitrary gray value) of eIF4A3 mRNA (± SEM; n=4-10/group) in dorsal striatum analyzed via radioactive in situ hybridization histochemistry and expressed as a percent of basal values in caged control (CC) animals. Rats in the CC group were sacrificed immediately upon removal from their home cage. Rats in the remaining groups explored a novel spatial environment (see Methods) for 5 min and were then either sacrificed immediately (“5 min” group) or returned to the home cage for 25 min before sacrifice (“30 min” group); or 55 min before sacrifice (“60 min” group). *Significantly different from CC (p=0.03). (B) Mean expression (arbitrary gray value) of Magoh mRNA (± SEM; n=4-7/group) in dorsal striatum analyzed via radioactive in situ hybridization histochemistry and expressed as a percent of basal values in CC animals. (C) A sense ribonucleotide probe for eIF4A3 gave no signal. (D) Striatal section from a CC rat labeled with the anti-sense ribonucleotide probe for eIF4A3.. (E) Striatal section from a “30 min” group rat labeled with the anti-sense ribonucleotide probe for eIF4A3.
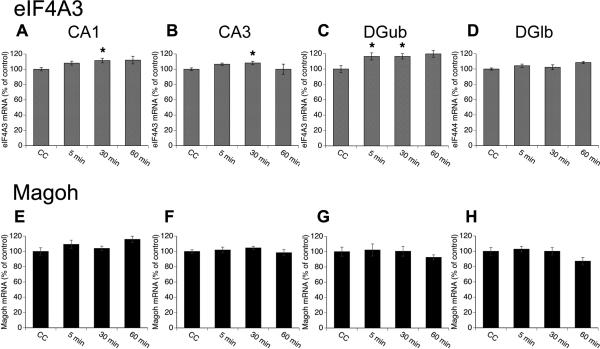
Given that Arc mRNA expression is increased in an activity-dependent manner in numerous brain regions (Guzowski et al., 1999, Chawla et al., 2005, Daberkow et al., 2007), we examined whether activity-dependent eIF4A3 mRNA expression was similarly broadly distributed in the brains of rats subsequent to spatial exploration of a novel environment. As in dorsal striatum, there was a time-dependent increase in eIF4A3 mRNA expression in dorsal hippocampus (Figure 2A-D), including the CA1 (“5 min”=108.2%±2.4, “30 min”=111.4%±3.0, “60 min”=128.5%±8.2; F(3,32)=3.18, p=0.04; Figure 2A) and CA3 (“5 min”=106.5%±1.5, “30 min”=108.3%±2.1, “60 min”=119.0%±5.8; F(3,32)=2.97, p=0.05; Figure 2B) subregions. Post-hoc analysis revealed significant increases in eIF4A3 mRNA expression over CC animals in the CA1 of the “30 min” group (p=0.02) and a trend for increases in expression over CC animals in the “60 min” group (p=0.08). Likewise, post-hoc analysis of the data from CA3 revealed a significant increase over CC animals in the “30 min” group (p=0.05). Similar to previous reports for novelty-induced Arc mRNA expression in the dentate gyrus (DG) subregion of hippocampus (Chawla et al., 2005), there was a time-dependent increase in eIF4A3 mRNA expression in the upper blade of the DG (DGub; F(3,32)=3.22, p=0.04). Post-hoc analysis revealed significant increases in eIF4A3 mRNA expression in the DGub of the “5 min” (116.4%±4.6, p=0.05) and “30 min” (116.5%±3.4, p=0.05; Figure 2C) groups relative to the CC group, as well as a trend for increase in expression in the “60 min” group (119.7%±3.4, p=0.07). Conversely, there was no change in eIF4A3 expression in the lower blade of the dentate gyrus (DGlb F(3,32)=1.37, p=0.3; Figure 2D) at any time point.
Figure 2. Expression of EJC components in dorsal hippocampus of rats engaged in spatial exploration for 5 min.
(A-D) Mean expression (arbitrary gray value) of eIF4A3 mRNA (± SEM; n=4-10/group) in the CA1 (A), CA3 (B), upper blade of the dentate gyrus (DGub; C), and lower blade of the dentate gyrus (DGlb; D) subregions of dorsal hippocampus analyzed via radioactive in situ hybridization histochemistry and expressed as a percent of basal values in caged control (CC) animals. *Significantly different from CC. (E-H) Mean expression (arbitrary gray value) of Magoh mRNA (± SEM; n=10/group) in CA1 (E), CA3 (F), DGub (G), and DGlb (H) analyzed via radioactive in situ hybridization histochemistry and expressed as a percent of basal values in CC animals.
As with striatum, there were no significant increases in Magoh mRNA expression in any regions of the hippocampus following spatial exploration (Figure 2E-H; CA1 “5 min”=109.9%±5.3, “30 min”=105.0±3.1, “60 min”=115.8%±4.1; F(3,31)=1.15, p>0.05; CA3 “5 min”=98.3%±4.2, “30 min”=103.0%±3.6, “60 min”=98.4%±4.1; F(3,31)=0.65, p>0.05; DGub “5 min”=103.4%±4.0, “30 min”=106.4%±1.8, “60 min”=92.6%±3.2; F(3,18)=0.35, p>0.05; DGlb “5 min”=96.8%±5.4, “30 min”=100.3±4.7, “60 min”=87.1%±5.0; F(3.18)=1.38, p>0.05). Thus, there were activity-related increases in eIF4A3, but not Magoh, mRNA in both dorsal striatum (Figure 1) and dorsal hippocampus (Figure 2). The activity-related expression of eIF4A3 is similar to the pattern of exploration-induced Arc mRNA expression previously reported, including time-dependent increases in both dorsal striatum (Daberkow et al., 2007) and CA1, CA3, and DGub subregions of hippocampus (Guzowski et al., 1999, Chawla et al., 2005, Vazdarjanova et al., 2006).
3.2. Activity-related increase in eIF4A3 protein distribution in striatum
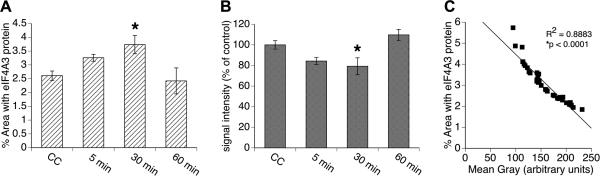
To further assess the activity-dependent regulation of eIF4A3, we used fluorescence immunohistochemistry to examine the expression of eIF4A3 protein after animals engaged in spatial exploration of a novel environment (Figure 3). CC animals exhibited low basal distribution of eIF4A3 protein (percent of total field area with signal) but high signal intensity (mean gray value of pixels). Following exploration, there was a significant time-dependent increase in the percent of the total field area with signal (F(4,37)=4.95, p=0.003; Fig. 3A), with the “30 min” group showing increased distribution of the eIF4A3 signal relative to the CC group (p<0.01). This distribution returned to basal levels by 60 minutes (Figure 3A), such that expression within the “60 min” group was not significantly different from the CC group (p>0.9). Furthermore, there was a significant time-dependent effect on eIF4A3 protein signal intensity (mean gray value) within striatum (F(4,37)=5.21, p=0.002; Figure 3B). Post-hoc analysis revealed a significant decrease in the signal intensity in the “30 min” group (p=0.02) relative to the CC group. As was the case for the distribution of the eIF4A3 signal, the increase in signal intensity returned to basal levels by 60 minutes, such that expression in the “60 min” group was not significantly different from the CC group (p=0.7). Correlating the measurement of percent field area with eIF4A3 signal to the signal intensity (mean gray value of pixels) revealed a strong inverse correlation (R2=0.89, p<0.0001; Figure 3C), suggesting that there is activity-dependent movement of this protein together with mRNA granules, such as Arc (Kanai et al., 2004), during periods of neuronal activation rather than de novo synthesis of protein
Figure 3. Expression of eIF4A3 protein in dorsal striatum of rats engaged in spatial exploration for 5 min.
(A) Mean (± SEM, n=4-10/group) eIF4A3 protein distribution in dorsal striatum measured as percent of total image area with signal (i.e. percentage of total pixel area in the field with eIF4A3-labeled pixels). *Significantly different from caged controls (CC; p<0.01). (B) Signal intensity (average gray area), expressed as mean percent of control (± SEM, n=4-10/group), of eIF4A3 protein-labeled pixels in dorsal striatum *Significantly different from CC (p=0.02). (C) Significant inverse correlation (p<0.05) between percent of total field with eIF4A3 protein signal above threshold and the average signal intensity (mean gray value) of the labeled.
3.3 Colocalization of eIF4A3 protein with Arc mRNA
To examine whether Arc mRNA and eIF4A3 protein interact in vivo, we performed double-label detection of Arc mRNA and eIF4A3 by combined FISH/fluorescent immunohistochemistry followed by confocal imaging in a subset of rats (n=3) following response-reversal learning on the T-maze (Figure 4). Areas of Arc mRNA and eIF4A3 protein colocalization are found throughout the dorsomedial striatum under these conditions (Figure 4A). To estimate the extent of colocalization, two separate image fields from each T-maze-trained rat were analyzed. On average, 43.5%±0.02 of Arc mRNA-positive puncta colocalized with eIF4A3 protein-positive puncta outside of the somatic compartment, whereas 33.9%±0.2 of eIF4A3 protein-positive puncta colocalized with Arc mRNA-positive puncta. A previous study reported that ~59% of Arc mRNA-positive puncta colocalized with eIF4A3 protein-puncta, whereas 29% of eIF4A3 protein-positive puncta colocalized with Arc mRNA-positive puncta in vitro following 6-hr incubation of cultured hippocampal neurons with BDNF (Giorgi et al., 2007), Additionally, another study reported that the neuron-specific RNA binding protein Smaug-1 also showed 40-60% colocalization with CaMKIIa mRNA following NMDA receptor stimulation in cultured hippocampal neurons in vitro, with this protein contributing to CaMKIIa mRNA availability and activity-dependent protein synthesis (Baez et al., 2011). Thus, our analysis of the in vivo colocalization of Arc and eIF4A3 following learning is in line with previous reports for RNA binding proteins colocalizing with target mRNAs following various pharmacological stimulation paradigms in vitro.
3.4. Correlation between eIF4A3 mRNA expression and striatally-mediated learning
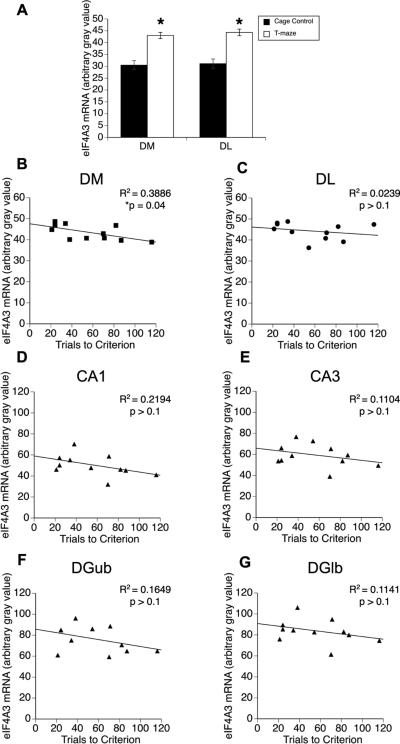
Previously, we reported that Arc mRNA expression in dorsomedial (DM), but not dorsolateral (DL), striatum of normal rats correlates with trials to criterion on a T-maze-based, response-reversal learning task (Daberkow et al., 2007). Furthermore, knockdown of Arc mRNA in DM striatum impairs consolidation of reversal learning on this task (Pastuzyn et al., 2012). Thus, we examined whether eIF4A3 mRNA expression correlates with behavioral performance. Similar to our prior observations with Arc mRNA (Daberkow et al., 2007, 2008), eIF4A3 mRNA expression significantly increased in DM and DL striatum of animals performing on the T-maze relative to CC rats (Figure 5A; CC - DM: 34.9±1.9; DL: 35.5±2.38; T-maze - DM: 43.3±1.1, t(10)=6.63, p=0.0001; DL: 44.0±1.2, t(10)=6.3, p=0.0001). However, only eIF4A3 mRNA expression in DM striatum (R2=0.39; p=0.04; Figure 5B) significantly correlated with behavioral performance (i.e. trials to criterion); eIF4A3 mRNA expression in DL striatum (R2=0.02; p>0.1; Figure 5C) or any subregion of dorsal hippocampus (CA1, R2=0.22; p>0.1; CA3, R2=0.11; p>0.1; DGub R2=0.16; p>0.1; DGlb R2=0.11; p>0.1; Figure 5D-E) did not correlate with behavioral performance.
Figure 5. Expression of eIF4A3 mRNA in the brains of rats undergoing response-reversal learning.
(A) Rats trained to perform on a response-reversal learning task on a T-maze showed significant increases in eIF4A3 mRNA expression in the dorsomedial (DM) and dorsolateral (DL) striatum compared to caged control (CC) rats (n = 11 per group). *Significantly different from CC (p=0.0001). (B-G) Degree of correlation between behavioral performance (trials to criterion) on the response-reversal task and eIF4A3 mRNA expression (average gray value from densitometric analysis) in DM striatum (A), DL striatum (B), CA1 of dorsal hippocampus (C), CA3 of dorsal hippocampus (D), upper blade of dentate gyrus (DGub; E), and lower blade of dentate gyrus (DGlb; F). *Significant correlation (p<0.05).
4. Discussion
We presently demonstrate increased expression of the EJC factor eIF4A3 in the adult rodent CNS under conditions of spatial exploration and striatally-mediated response-reversal learning. The level of eIF4A3, but interestingly not Magoh, mRNA increased in an activation-related manner across multiple brain regions, and expression of eIF4A3 mRNA in DM striatum correlated with behavioral performance on a striatally-based response-reversal learning task. Furthermore, eIF4A3 protein colocalized with Arc mRNA in vivo following striatally-mediated learning. Given the complex signaling required of neurons undergoing activity-dependent synaptic modifications, it is conceivable that such plasticity requires activity-dependent expression of eIF4A3 to facilitate normal mRNA quality control through NMD (Chang et al., 2007, Shyu et al., 2008) and to regulate the translation and decay of plasticity-related mRNAs, such as Arc (Giorgi et al., 2007, Soule et al., 2012). The present observations are the first to demonstrate activity-dependent changes in eIF4A3 mRNA and protein levels in the adult mammalian brain in vivo, further implicating eIFAA3 as a mediator of activity-dependent neuroplasticity processes.
eIF4A3 likely plays an important role in neuronal mRNA processing given its core role in EJC formation and function (Shibuya et al., 2004, Ballut et al., 2005). Numerous aspects of post-transcriptional mRNA processing are mediated by the EJC, including nucleocytoplasmic shuttling (Shibuya et al., 2004), cytoplasmic translational control (Diem et al., 2007), and NMD/TDD (Maquat, 2004). Specifically, continued presence of the EJC on spliced mRNAs following the first round of protein translation can initiate mRNA decay by NMD (Maquat, 2004, Shibuya et al., 2004), thereby tightly limiting mRNA availability. Given the exclusive role that eIF4A3 plays in EJC formation (Chan et al., 2004, Shibuya et al., 2004) and NMD (Ferraiuolo et al., 2004), it is likely a critical component of mRNA stability underlying normal neuronal signaling in the adult mammalian CNS.
Numerous neuronal mRNAs are dendritically targeted for local protein synthesis, indicating a need for tight post-transcriptional and translational regulation of these mRNAs to coordinate neuronal signaling (Ule and Darnell, 2006, Goldie and Cairns, 2012). The EJC, and eIF4A3 specifically, may thus allow neurons to regulate the expression of specific effector mRNAs, such as Arc, through NMD/TDD or through other post-transcriptional processes involved in the trafficking of such effector mRNAs to or translational control of those mRNAs at synapses. Unlike other eIF4A isoforms (Li et al., 1999), which are components of the eIF4F translation initiation complex (Klann and Dever, 2004), eIF4A3 plays a distinct role in mRNA stability. Understanding whether these other eIF4A isoforms also show activity-dependent changes in expression will help clarify the precise contribution made by the EJC and NMD vs. translational regulation in determining synaptic plasticity processes. However, given the known role of eIF4A3 in mRNA regulation and its activity-related expression, mRNA stability processes mediated by NMD may act as a potential means by which neurons can regulate response to synaptic stimuli.
As demonstrated by others (Giorgi et al., 2007, Soule et al., 2012) and now us (Figure 4), one potential means for eIF4A3 to contribute to synaptic plasticity is through interaction with and regulation of Arc mRNA. As eIF4A3 is critical to the EJC (Chan et al., 2004, Shibuya et al., 2004, Ballut et al., 2005), it may act as a brake on cytoplasmic Arc mRNA availability through NMD (Giorgi et al., 2007). Arc is a target for NMD due to the two EJCs within the 3’UTR, leading to tight control of Arc protein synthesis (Giorgi et al., 2007, Soule et al., 2012). Knockdown of eIF4A3 increased Arc mRNA and protein levels in rat hippocampal somata and dendrites in vitro and increased mEPSC amplitude and synaptic mGLUR1 levels (Giorgi et al., 2007), indicating that eIF4A3 can directly affect Arc-dependent synaptic plasticity. This present work demonstrates time-dependent changes in eIF4A3 protein distribution and signal intensity following exploration of a novel environment. Additionally, eIF4A3 protein colocalized with Arc mRNA in the dorsal striatum following striatally-mediated learning (Figure 4), similar to previous reports in vitro (Giorgi et al., 2007). Thus, one potential explanation for the observed increase in distribution of eIF4A3 protein is that neuronal activation associated with spatial exploration or engagement on a learning and memory task induces de novo transcription of effector mRNAs, such as Arc (Guzowski et al., 1999, Daberkow et al., 2007), which in turn leads to the formation of eIF4A3-containing protein-mRNA granules (Kanai et al., 2004) that then distribute out into dendrites. Such greater distribution of the eIF4A3-containing granules throughout the neuropil may then result in the lower signal intensity in any given set of labeled pixels. However, we cannot presently rule out the possibility that the observed changes in eIF4A3 protein expression also reflect de novo eIF4A3 protein synthesis. Many plasticity-associated mRNAs, including CaMKIIα (Ouyang et al., 1997), zif268 (Zangenehpour and Chaudhuri, 2002) and even Arc (Vazdarjanova et al., 2006, Niere et al., 2012), demonstrate rapid protein synthesis in response to stimuli. Thus, while it was somewhat surprising that the percent of the total field area with eIF4A3 protein increased relatively rapidly in response to exploration of a novel environment, this timeframe is not unrealistic for neuronal protein translation and would thus supply an alternative interpretation of the presently observed changes in eIF4A3 protein expression
Our novel findings for eIF4A3 mRNA and protein levels parallel those previously reported for Arc mRNA (Guzowski et al., 1999, Chawla et al., 2005, Daberkow et al., 2007). First, eIF4A3 mRNA is rapidly increased in dorsal striatum (Daberkow et al., 2007), as well as in CA1, CA3, and DGub of dorsal hippocampus (Guzowski et al., 1999) of adult rats engaged in brief (5 min) exploration of a novel environment. Second, like Arc mRNA (Chawla et al., 2005, Vazdarjanova et al., 2006), eIF4A3 mRNA expression was not increased in DGlb. Third, there is a significant correlation between eIF4A3 mRNA levels in DM striatum, but not DL striatum or dorsal hippocampal subfields, and trials to criterion on a striatally-based, response-reversal learning task (Daberkow et al., 2007, 2008). Fourth, Arc mRNA colocalized with eIF4A3 protein in vivo following striatally-based learning, as it has been shown to do in the hippocampus in vitro as well (Giorgi et al., 2007). Finally, there was a significant activity-dependent increase in eIF4A3 protein distribution that returned to basal levels 60 min after exploration of a novel environment, in close parallel to the time frame of Arc transcription, trafficking, and protein translation (Vazdarjanova et al., 2006, Baez et al., 2011). These findings suggest that eIF4A3 may critically regulate Arc-dependent synaptic plasticity. In this regard, it is interesting that EJC components also regulate MAP kinase (MAPK) splicing in Drosophila (Ashton-Beaucage et al., 2010, Roignant and Treisman, 2010). MAPK is critical for ERK phosphorylation, and thus, Arc mRNA targeting to activated synapses (Huang et al., 2007, Wang et al., 2009). The in vivo observations reported herein are the first to demonstrate activity-related regulation of the core EJC component eIF4A3, with the expression of eIF4A3 mRNA showing notable similarities to the behaviorally-induced expression of Arc mRNA previously reported by others (Guzowski et al., 1999, Chawla et al., 2005, Vazdarjanova et al., 2006) and us (Daberkow et al., 2007, 2008, Pastuzyn et al., 2012). The extent to which eIF4A3 is necessary for Arc-dependent neuroplasticity in the brain in vivo is currently under further investigation.
Presently, the basis for the activity-related expression of eIF4A3 mRNA levels is unknown, but the current findings further highlight that neurons dynamically regulate mRNAs involved in post-transcriptional processing, such as RNA binding. For example, the mediator of cap-binding activity during protein translation initiation, eIF4E mRNA, shows dendritic localization and increased association with PSD-95 upon KCl stimulation in vitro (Moon et al., 2009), demonstrating that neurons not only dendritically localize effector genes, but also the factors that facilitate local translation of those effector genes. Additionally, the brain-specific, ELAV/Hu family of RNA-binding proteins, which are known to contribute to GAP-43 mRNA localization and cytoplasmic stabilization, also show dendritic localization (Bolognani et al., 2004) and activity-dependent expression following spatial learning (Pascale et al., 2004). Furthermore, ELAV/Hu expression is modulated by glutamatergic (seizures) or dopaminergic (cocaine) signaling (Tiruchinapalli et al., 2008), thereby demonstrating a role of classical neurotransmitter systems converging onto activity-dependent, post-transcriptional regulatory elements to modulate neuronal responses. Thus, future work will clarify whether neuronal stimulation results in new transcriptional activation of eIF4A3 or alternative post-transcriptional processing of eIF4A3, so as to determine the basis for the observed activity-dependent changes in eIF4A3 mRNA and protein levels.
5. Conclusions
Our present findings reveal activity-related increases in eIF4A3 mRNA and protein distribution in the adult mammalian CNS in vivo. Previous reports indicate a unique function for eIF4A3 in regulating expression of Arc mRNA (Giorgi et al., 2007), a key mediator of synaptic plasticity (Guzowski et al., 2000, Chowdhury et al., 2006, Rial Verde et al., 2006) and basal ganglia-mediated learning consolidation (Pastuzyn et al., 2012). Herein, we demonstrate that eIF4A3, but not Magoh, mRNA shows striking similarities to Arc mRNA in terms of patterns of expression in the context of spatial exploration (Guzowski et al., 1999, Chawla et al., 2005) and the correlation of mRNA expression with learning (Daberkow et al., 2007, 2008), implicating eIF4A3 as a potential regulator of Arc expression in vivo. These present observations suggest that neurons coordinate the expression of not only effector genes, such as Arc, but also post-transcriptional regulatory factors and pathways required by those specific effector genes. Dysfunction in these factors and pathways potentially disrupts neuroplasticity processes and can lead to neurological disorders (Dahm and Macchi, 2007, Tarpey et al., 2007). However, how these regulatory elements are themselves expressed and behaviorally activated in the adult, mammalian brain is currently less clear. Future studies are thus needed to determine the precise mechanisms by which eIF4A3 is regulated in vivo, as well as the functional roles that eIF4A3 may be playing in the post-transcriptional regulation of select effector genes within activated neurons. Such knowledge will afford a more complete understanding of both normal and abnormal neuroplasticity processes required of the adult mammalian CNS.
Highlights.
Exon junction complex components eIF4A3 and Magoh expression investigated in vivo.
Activity-dependent expression of eIF4A3 parallels that previously reported for Arc.
eIF4A3 expression increased in striatum and hippocampus with behavioral activation.
Striatal eIF4A3 mRNA expression correlates with striatally-based learning.
Neurons may dynamically control mRNA regulatory elements needed for plasticity.
Acknowledgements
This work was supported by NIH grant DA024036 (KAK); American Foundation for Pharmaceutical Education (MBH), Parkinson's Disease Foundation (MBH); NIDA DA032502 (EDP). The authors wish to thank Ms. Bethany Pelton for assistance with preliminary hippocampal data analysis.
Abbreviations
- eIF4A3
eukaryotic initiation factor 4A3
- EJC
exon-junction complex
- Arc/Arg3.1
activity-regulated, cytoskeleton-associated protein
- TDD
translation dependent decay
- NMD
nonsense-mediated mRNA decay
- CC
cage control
- CNS
central nervous system
- PBS-T
PBS/0.1% Triton-X
- PBS-TB
PBS/0.1% Triton-X/0.1% Bovine Serum Albumin
- TNT
TBS/0.05% Tween-20
- TSA
tyramide signal amplification
- DIG-UTP
digoxigenin-conjugated UTP
- FISH
fluorescence in situ hybridization
- DM
dorsomedial
- DL
dorsolateral
- DGub
upper blade of dentate gyrus
- DGlb
lower blade of dentate gyrus
- FMRP
Fragile X mental retardation protein
- BDNF
brain-derived neurotrophic factor
- HRP
horseradish peroxidase
- mGLUR1
metabotropic glutamate receptor 1
- mEPSC
miniature excitatory postsynaptic current
- ELAV
embryonic lethal, abnormal vision protein
- GAP-43
growth associated protein 43
- MAPK
mitogen-activated protein kinase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of Interests: None
References
- Ashton-Beaucage D, Udell CM, Lavoie H, Baril C, Lefrancois M, Chagnon P, Gendron P, Caron-Lizotte O, Bonneil E, Thibault P, Therrien M. The exon junction complex controls the splicing of MAPK and other long intron-containing transcripts in Drosophila. Cell. 2010;143:251–262. doi: 10.1016/j.cell.2010.09.014. [DOI] [PubMed] [Google Scholar]
- Baez MV, Luchelli L, Maschi D, Habif M, Pascual M, Thomas MG, Boccaccio GL. Smaug1 mRNA-silencing foci respond to NMDA and modulate synapse formation. J Cell Biol. 2011;195:1141–1157. doi: 10.1083/jcb.201108159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballut L, Marchadier B, Baguet A, Tomasetto C, Seraphin B, Le Hir H. The exon junction core complex is locked onto RNA by inhibition of eIF4AIII ATPase activity. Nat Struct Mol Biol. 2005;12:861–869. doi: 10.1038/nsmb990. [DOI] [PubMed] [Google Scholar]
- Bolognani F, Merhege MA, Twiss J, Perrone-Bizzozero NI. Dendritic localization of the RNA-binding protein HuD in hippocampal neurons: association with polysomes and upregulation during contextual learning. Neurosci Lett. 2004;371:152–157. doi: 10.1016/j.neulet.2004.08.074. [DOI] [PubMed] [Google Scholar]
- Chan CC, Dostie J, Diem MD, Feng W, Mann M, Rappsilber J, Dreyfuss G. eIF4A3 is a novel component of the exon junction complex. RNA. 2004;10:200–209. doi: 10.1261/rna.5230104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YF, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- Chawla MK, Guzowski JF, Ramirez-Amaya V, Lipa P, Hoffman KL, Marriott LK, Worley PF, McNaughton BL, Barnes CA. Sparse, environmentally selective expression of Arc RNA in the upper blade of the rodent fascia dentata by brief spatial experience. Hippocampus. 2005;15:579–586. doi: 10.1002/hipo.20091. [DOI] [PubMed] [Google Scholar]
- Chowdhury S, Shepherd JD, Okuno H, Lyford G, Petralia RS, Plath N, Kuhl D, Huganir RL, Worley PF. Arc/Arg3.1 interacts with the endocytic machinery to regulate AMPA receptor trafficking. Neuron. 2006;52:445–459. doi: 10.1016/j.neuron.2006.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daberkow DP, Riedy MD, Kesner RP, Keefe KA. Arc mRNA induction in striatal efferent neurons associated with response learning. Eur J Neurosci. 2007;26:228–241. doi: 10.1111/j.1460-9568.2007.05630.x. [DOI] [PubMed] [Google Scholar]
- Daberkow DP, Riedy MD, Kesner RP, Keefe KA. Effect of methamphetamine neurotoxicity on learning-induced Arc mRNA expression in identified striatal efferent neurons. Neurotox Res. 2008;14:307–315. doi: 10.1007/BF03033855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahm R, Macchi P. Human pathologies associated with defective RNA transport and localization in the nervous system. Biol Cell. 2007;99:649–661. doi: 10.1042/BC20070045. [DOI] [PubMed] [Google Scholar]
- Diem MD, Chan CC, Younis I, Dreyfuss G. PYM binds the cytoplasmic exon-junction complex and ribosomes to enhance translation of spliced mRNAs. Nat Struct Mol Biol. 2007;14:1173–1179. doi: 10.1038/nsmb1321. [DOI] [PubMed] [Google Scholar]
- Dostie J, Dreyfuss G. Translation is required to remove Y14 from mRNAs in the cytoplasm. Curr Biol. 2002;12:1060–1067. doi: 10.1016/s0960-9822(02)00902-8. [DOI] [PubMed] [Google Scholar]
- Ferraiuolo MA, Lee CS, Ler LW, Hsu JL, Costa-Mattioli M, Luo MJ, Reed R, Sonenberg N. A nuclear translation-like factor eIF4AIII is recruited to the mRNA during splicing and functions in nonsense-mediated decay. Proc Natl Acad Sci U S A. 2004;101:4118–4123. doi: 10.1073/pnas.0400933101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguly A, Keefe KA. Unilateral dopamine depletion increases expression of the 2A subunit of the N-methyl-D-aspartate receptor in enkephalin-positive and enkephalin-negative neurons. Neuroscience. 2001;103:405–412. doi: 10.1016/s0306-4522(01)00005-7. [DOI] [PubMed] [Google Scholar]
- Giorgi C, Yeo GW, Stone ME, Katz DB, Burge C, Turrigiano G, Moore MJ. The EJC factor eIF4AIII modulates synaptic strength and neuronal protein expression. Cell. 2007;130:179–191. doi: 10.1016/j.cell.2007.05.028. [DOI] [PubMed] [Google Scholar]
- Goldie BJ, Cairns MJ. Post-transcriptional trafficking and regulation of neuronal gene expression. Mol Neurobiol. 2012;45:99–108. doi: 10.1007/s12035-011-8222-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzowski JF, Lyford GL, Stevenson GD, Houston FP, McGaugh JL, Worley PF, Barnes CA. Inhibition of activity-dependent arc protein expression in the rat hippocampus impairs the maintenance of long-term potentiation and the consolidation of long-term memory. J Neurosci. 2000;20:3993–4001. doi: 10.1523/JNEUROSCI.20-11-03993.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzowski JF, McNaughton BL, Barnes CA, Worley PF. Environment-specific expression of the immediate-early gene Arc in hippocampal neuronal ensembles. Nat Neurosci. 1999;2:1120–1124. doi: 10.1038/16046. [DOI] [PubMed] [Google Scholar]
- Huang F, Chotiner JK, Steward O. Actin polymerization and ERK phosphorylation are required for Arc/Arg3.1 mRNA targeting to activated synaptic sites on dendrites. J Neurosci. 2007;27:9054–9067. doi: 10.1523/JNEUROSCI.2410-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai Y, Dohmae N, Hirokawa N. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron. 2004;43:513–525. doi: 10.1016/j.neuron.2004.07.022. [DOI] [PubMed] [Google Scholar]
- Klann E, Dever TE. Biochemical mechanisms for translational regulation in synaptic plasticity. Nat Rev Neurosci. 2004;5:931–942. doi: 10.1038/nrn1557. [DOI] [PubMed] [Google Scholar]
- Li Q, Imataka H, Morino S, Rogers GW, Jr., Richter-Cook NJ, Merrick WC, Sonenberg N. Eukaryotic translation initiation factor 4AIII (eIF4AIII) is functionally distinct from eIF4AI and eIF4AII. Mol Cell Biol. 1999;19:7336–7346. doi: 10.1128/mcb.19.11.7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maquat LE. Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nat Rev Mol Cell Biol. 2004;5:89–99. doi: 10.1038/nrm1310. [DOI] [PubMed] [Google Scholar]
- Moon IS, Cho SJ, Seog DH, Walikonis R. Neuronal activation increases the density of eukaryotic translation initiation factor 4E mRNA clusters in dendrites of cultured hippocampal neurons. Exp Mol Med. 2009;41:601–610. doi: 10.3858/emm.2009.41.8.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niere F, Wilkerson JR, Huber KM. Evidence for a fragile X mental retardation protein-mediated translational switch in metabotropic glutamate receptor-triggered Arc translation and long-term depression. J Neurosci. 2012;32:5924–5936. doi: 10.1523/JNEUROSCI.4650-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang Y, Kantor D, Harris KM, Schuman EM, Kennedy MB. Visualization of the distribution of autophosphorylated calcium/calmodulin-dependent protein kinase II after tetanic stimulation in the CA1 area of the hippocampus. J Neurosci. 1997;17:5416–5427. doi: 10.1523/JNEUROSCI.17-14-05416.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panja D, Dagyte G, Bidinosti M, Wibrand K, Kristiansen AM, Sonenberg N, Bramham CR. Novel translational control in Arc-dependent long term potentiation consolidation in vivo. J Biol Chem. 2009;284:31498–31511. doi: 10.1074/jbc.M109.056077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascale A, Gusev PA, Amadio M, Dottorini T, Govoni S, Alkon DL, Quattrone A. Increase of the RNA-binding protein HuD and posttranscriptional up-regulation of the GAP-43 gene during spatial memory. Proc Natl Acad Sci U S A. 2004;101:1217–1222. doi: 10.1073/pnas.0307674100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastuzyn ED, Chapman DE, Wilcox KS, Keefe KA. Altered learning and arc-regulated consolidation of learning in striatum by methamphetamine-induced neurotoxicity. Neuropsychopharmacology. 2012;37:885–895. doi: 10.1038/npp.2011.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rial Verde EM, Lee-Osbourne J, Worley PF, Malinow R, Cline HT. Increased expression of the immediate-early gene arc/arg3.1 reduces AMPA receptor-mediated synaptic transmission. Neuron. 2006;52:461–474. doi: 10.1016/j.neuron.2006.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roignant JY, Treisman JE. Exon junction complex subunits are required to splice Drosophila MAP kinase, a large heterochromatic gene. Cell. 2010;143:238–250. doi: 10.1016/j.cell.2010.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibuya T, Tange TO, Sonenberg N, Moore MJ. eIF4AIII binds spliced mRNA in the exon junction complex and is essential for nonsense-mediated decay. Nat Struct Mol Biol. 2004;11:346–351. doi: 10.1038/nsmb750. [DOI] [PubMed] [Google Scholar]
- Shyu AB, Wilkinson MF, van Hoof A. Messenger RNA regulation: to translate or to degrade. EMBO J. 2008;27:471–481. doi: 10.1038/sj.emboj.7601977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soule J, Alme M, Myrum C, Schubert M, Kanhema T, Bramham CR. Balancing Arc Synthesis, mRNA Decay, and Proteasomal Degradation: Maximal Protein Expression Triggered by Rapid Eye Movement Sleep-Like Bursts of Muscarinic Cholinergic Recpetor Stimulation. J Biol Chem. 2012;287:22354–22366. doi: 10.1074/jbc.M112.376491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton MA, Schuman EM. Dendritic protein synthesis, synaptic plasticity, and memory. Cell. 2006;127:49–58. doi: 10.1016/j.cell.2006.09.014. [DOI] [PubMed] [Google Scholar]
- Tange TO, Nott A, Moore MJ. The ever-increasing complexities of the exon junction complex. Curr Opin Cell Biol. 2004;16:279–284. doi: 10.1016/j.ceb.2004.03.012. [DOI] [PubMed] [Google Scholar]
- Tange TO, Shibuya T, Jurica MS, Moore MJ. Biochemical analysis of the EJC reveals two new factors and a stable tetrameric protein core. RNA. 2005;11:1869–1883. doi: 10.1261/rna.2155905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarpey PS, Raymond FL, Nguyen LS, Rodriguez J, Hackett A, Vandeleur L, Smith R, Shoubridge C, Edkins S, Stevens C, O'Meara S, Tofts C, Barthorpe S, Buck G, Cole J, Halliday K, Hills K, Jones D, Mironenko T, Perry J, Varian J, West S, Widaa S, Teague J, Dicks E, Butler A, Menzies A, Richardson D, Jenkinson A, Shepherd R, Raine K, Moon J, Luo Y, Parnau J, Bhat SS, Gardner A, Corbett M, Brooks D, Thomas P, Parkinson-Lawrence E, Porteous ME, Warner JP, Sanderson T, Pearson P, Simensen RJ, Skinner C, Hoganson G, Superneau D, Wooster R, Bobrow M, Turner G, Stevenson RE, Schwartz CE, Futreal PA, Srivastava AK, Stratton MR, Gecz J. Mutations in UPF3B, a member of the nonsense-mediated mRNA decay complex, cause syndromic and nonsyndromic mental retardation. Nat Genet. 2007;39:1127–1133. doi: 10.1038/ng2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiruchinapalli DM, Caron MG, Keene JD. Activity-dependent expression of ELAV/Hu RBPs and neuronal mRNAs in seizure and cocaine brain. J Neurochem. 2008;107:1529–1543. doi: 10.1111/j.1471-4159.2008.05718.x. [DOI] [PubMed] [Google Scholar]
- Ule J, Darnell RB. RNA binding proteins and the regulation of neuronal synaptic plasticity. Curr Opin Neurobiol. 2006;16:102–110. doi: 10.1016/j.conb.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Vazdarjanova A, Ramirez-Amaya V, Insel N, Plummer TK, Rosi S, Chowdhury S, Mikhael D, Worley PF, Guzowski JF, Barnes CA. Spatial exploration induces ARC, a plasticity-related immediate-early gene, only in calcium/calmodulin-dependent protein kinase II- positive principal excitatory and inhibitory neurons of the rat forebrain. J Comp Neurol. 2006;498:317–329. doi: 10.1002/cne.21003. [DOI] [PubMed] [Google Scholar]
- Wang Y, Zheng F, Zhou X, Sun Z, Wang H. Converging signal on ERK1/2 activity regulates group I mGluR-mediated Arc transcription. Neurosci Lett. 2009;460:36–40. doi: 10.1016/j.neulet.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zangenehpour S, Chaudhuri A. Differential induction and decay curves of c-fos and zif268 revealed through dual activity maps. Brain Res Mol Brain Res. 2002;109:221–225. doi: 10.1016/s0169-328x(02)00556-9. [DOI] [PubMed] [Google Scholar]