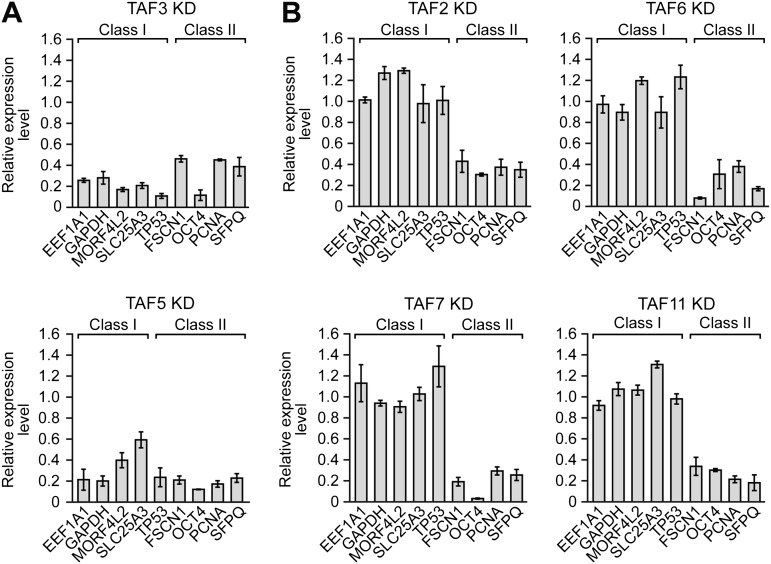
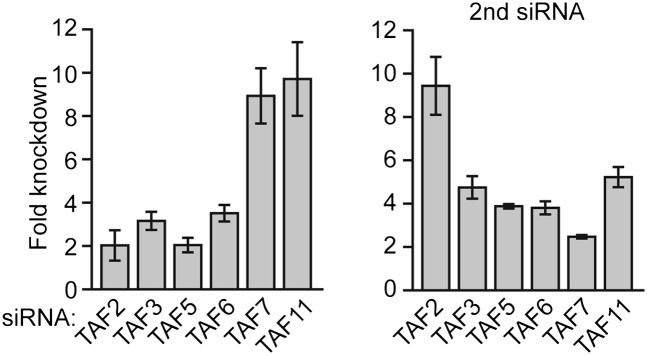
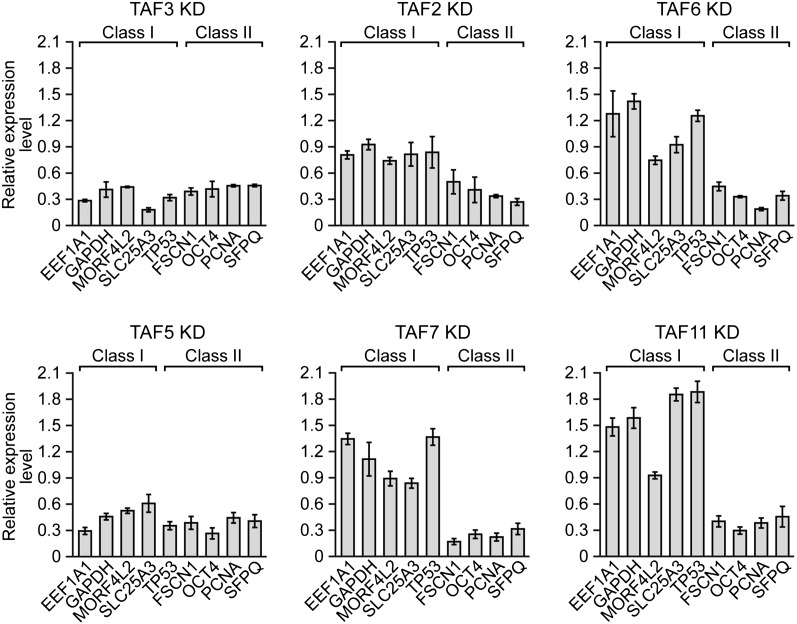
Figure 5. A strong relationship between TAF occupancy and transcriptional activity in hESCs.
(A) qRT-PCR analysis monitoring expression of class I and II genes in H9 TAF3 or TAF5 knockdown (KD) cells. Normalized Ct values were analyzed after subtracting the signal obtained with the control RN18S1 shRNA (see ‘Materials and methods’). Data are represented as mean ± SEM. (B) qRT-PCR analysis monitoring expression of class I and II genes in H9 TAF2, 6, 7, or 11 KD cells. Data are represented as mean ± SEM.