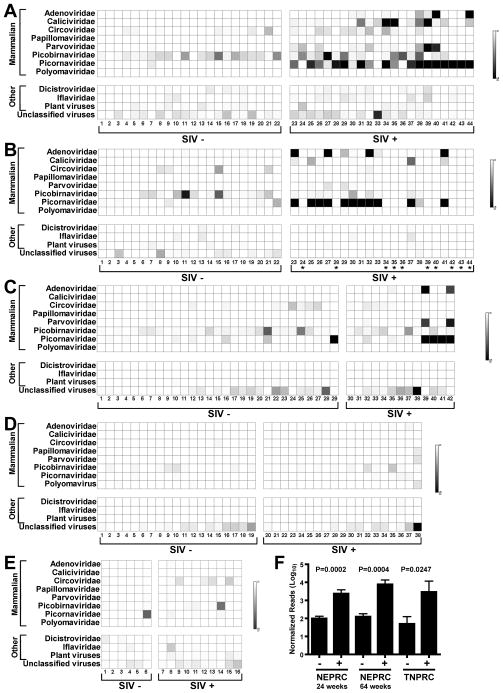
Figure 2. Distribution of virus sequences in rhesus and African green monkeys.
For these charts ‘mammalian’ indicates that sequences were most closely related to viruses that infect mammals. Viruses infecting non-mammals are referred to as ‘other’. ‘Unclassified virus’ includes all unclassified viruses, e.g. Chronic bee paralysis virus, Chimpanzee stool associated circular ssDNA virus, Circovirus-like genome RW-C, Circovirus-like genome CB-A, Rodent stool-associated circular genome virus, etc.
(A–E) The numbers below each chart and SIV infection status and panels are as in the Figure 1 legend.
(F) Comparison of the mean +/− SEM number of picornavirus sequences, after normalization for analysis using MEGAN, detected in the indicated cohorts of SIV-infected (+) and control (−) rhesus monkeys.
See also Table S1