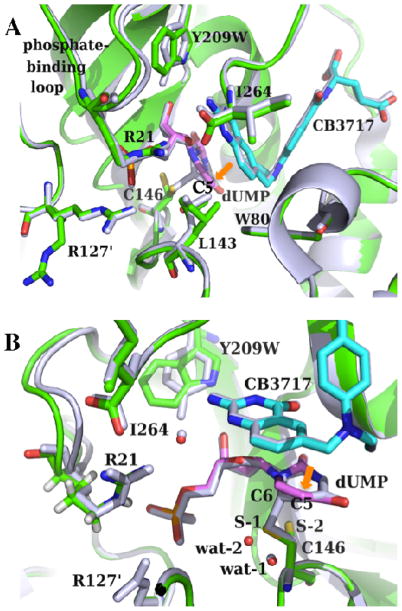
Figure 3.
(A) Crystal structures of the ternary complexes of the WT (gray; PDB ID 2G8O) and Y209W (green; PDB ID 4GEV) ecTSase with dUMP (magenta) and CB3717 (cyan) are remarkably similar. The mutant shows two conformations for each of the residues R127′, L143, and C146. The mutation also causes interesting dynamic effect including the shift-away and increased mobility of the phosphate-binding loop, and the less correlated atomic vibrations of protein residues and ligands. All the catalyzed chemical bond activations happen around the C5 of dUMP (orange arrow). (B) A closer view of the active site. The additional conformation of C146 (S-2) in Y209W cannot form a covalent bond with dUMP, which is accompanied by the displacement of a nearby water molecule (from wat-1 to wat-2). R21 on this phosphate-binding loop also forms H-bonds with both I264 and the cofactor (through a water molecule).