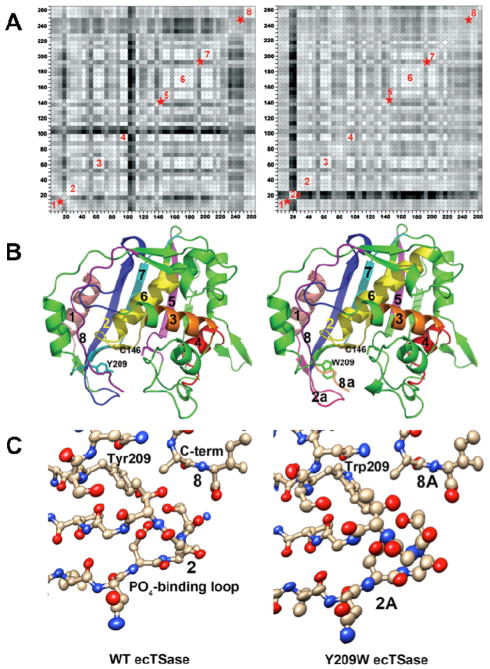
Figure 4.
(A) Plots showing correlations of anisotropic B-factor displacements between ecTSase atom pairs identified by the (x,y) grid coordinates of the plot (reproduced from Ref 41 with permission form ACS). The left plot is for WT ecTSase and the right plot is for Y209W. Grid points are shaded from white to black with white representing highest correlation, where degree of correlation is determined by similarity of projections of anisotropic displacements along the interatomic vector.59 Blocks of light colored squares along the diagonals of the plots indicate protein segments that vibrate as rigid bodies. Eight such segments are labeled in the plots, and red stars indicate segments where rigid body movement appears to have been disrupted by the mutation. Segment 2a in the Y209W plot is the phosphate-binding loop, whose vibrations are no longer correlated with the rest of the proteins segments. (B) Ribbon plots of WT ecTSase (left) and Y209W ecTSase (right). Segments labeled in (A) are labeled and colored. The catalytic cysteine and the mutated residue are shown as sticks. (C) Plot of the thermal ellipsoids of the phosphate-binding loop for WT (left) and Y209W ecTSase (right). This loop shifts to close the active site cavity during binding of the substrate and cofactor, and it has a key role in orienting the ligands and shielding the cavity from bulk solvent. R21 in this loop makes hydrogen bonds to the phosphate moiety of dUMP and to the protein C-terminus. The thermal ellipsoids are derived from the refined anisotropic B-factors of the structures and the radii of the ellipsoids are proportional to the root-mean-square displacements from the atom positions (see Experimental Details). The B-factors for the loop are larger in the mutant structure, as seen by the larger volumes of the ellipsoids. The thermal vibrations of atoms in the loop are also less well correlated with each other in the mutant, as evidenced by more random orientations of the long axes of the ellipsoids. In addition, the thermal vibrations of the atoms in the loop are no longer correlated with those of other protein segments lining the active site cavity, such as the segments containing S210 and the mutated Y209 or the C-terminal segment. The phosphate-binding loops and C-terminal residues are labeled with the segment numbers from the plots in (A).