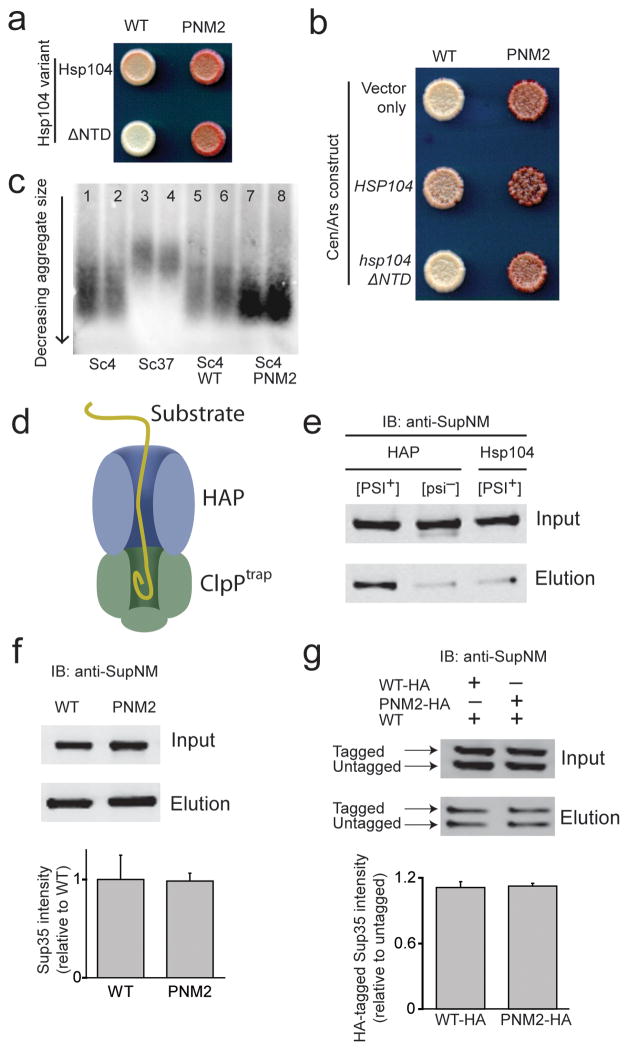
Figure 5.
PNM2 fibers in the Sc4 conformation interact with the in vivo chaperone machinery. (a) HSP104 (top row) and hsp104ΔNTD (bottom row) were expressed in [PSI+]Sc4 yeast expressing either WT (left) or PNM2 (right) Sup35p and spotted onto low adenine media. (b) HSP104 and hsp104ΔNTD were expressed at increased levels along with endogenous Hsp104p in [PSI+]Sc4 yeast expressing WT or PNM2 Sup35p. “Vector only” denotes transformation with an empty CEN/ARS plasmid. (c) SDD-AGE analysis of prion particle size in duplicate from lysates of [PSI+]Sc4 (lanes 1–2 and 5–6), [PSI+]Sc37 (lanes 3–4) and PNM2 [PSI+]Sc4 (lanes 7–8). SUP35 is genomic in lanes 1–4 and on a plasmid in lanes 5–8. Yeast express wild type HSP104 in all lanes. (d) Schematic of the in vivo HAP/ClpPtrap reaction. (e) Representative blot of the ClpPtrap affinity purification. WT Sup35p and ClpPtrap were expressed in backgrounds that were [PSI+]Sc4 or [psi−] expressing HAP or Hsp104p. (f) Monitoring the translocation of PNM2. HAP and ClpPtrap were expressed in [PSI+]Sc4 cells that also expressed WT or PNM2 Sup35p. Intensities of Sup35p elution signal from western blot (top panel) were normalized for input signal (bottom panel). (g) Monitoring translocation of PNM2 when co-expressed with WT. As in (f), but untagged WT Sup35p was also expressed in cells expressing HA-tagged WT or PNM2 Sup35p. The higher molecular weight band corresponds to HA-tagged Sup35p. Intensities of tagged-Sup35p elution signals were normalized for input signal and for untagged-Sup35p elution signal. For Sup35 intensity, values are expressed as mean ± s.e.m for three experiments.