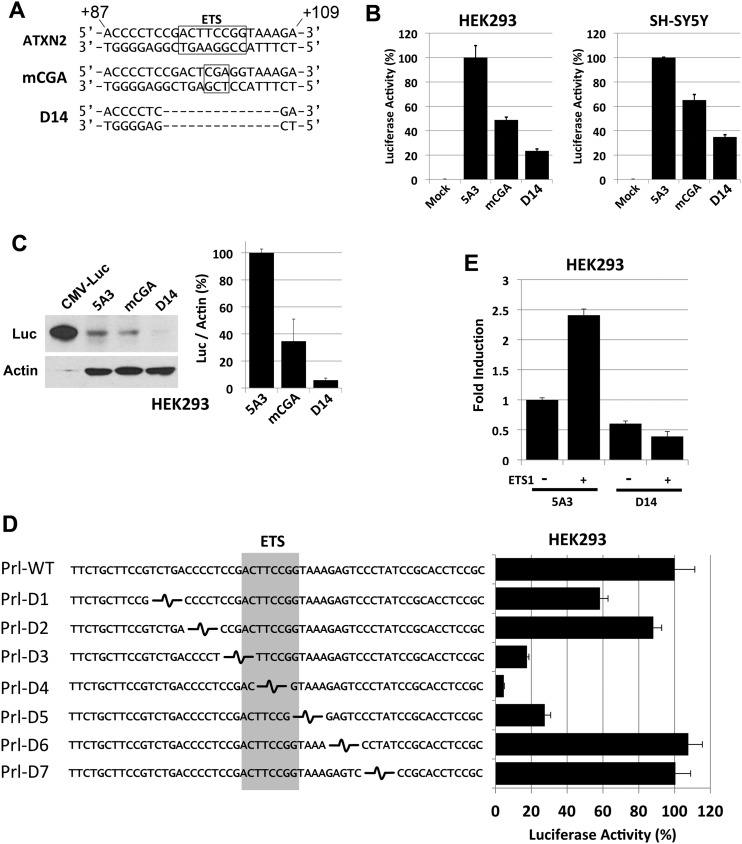
Figure 5.
A 14 bp region within the +87 to +109 ATXN2 5′-UTR region contains an ETS TF-binding site and accounts for the majority of ATXN2 expression. (A) Both strands of the ATXN2 sequence from +87 to +109 are shown and the position of an ETS TF-binding site oriented from +103→+96 on the ATXN2 minus strand is indicated (box). mCGA represents a TCC→CGA mutation with the ETS TF-binding site made in the ATXN2-luc expression plasmid pGL2-5A3. This mutation alters the ETS TF consensus sequence core on the ATXN2 minus strand from GGAA→TCGA to abrogate binding by ETS TFs. D14 represents a 14 bp deletion spanning the ETS TF-binding site in pGL2-5A3. (B) Luciferase assays comparing no mutation to the mCGA and D14 mutations in HEK293 and SH-SY5Y cells. The mCGA mutation resulted in 51 and 35% reduction in the average ATXN2-luc expression in HEK293 and SH-SY5Y cells, respectively, relative to no mutation. The D14 mutation resulted in 76 and 65% reduction in the average ATXN2-luc expression in HEK293 and SH-SY5Y cells, respectively, relative to no mutation. Values are means and SD from three independent transfections each read in triplicate and expression was controlled with SV40-Renilla luciferase. (C) Western blot showing that the mCGA and D14 mutations in pGL2-5A3 reduced ATXN2-luc expression in HEK293 cells compared with no mutation. The D14 mutation reduced ATXN2-luc to background. (D) Effect of seven tandem 5 bp deletions in the ATXN2 promoter on ATXN2-luc expression. Deletions were made by ligating annealed oligonucleotides spanning 58 bp of the ATXN2 promoter into a rat prolactin-luc minimal promoter construct. Deletions overlapping the ETS element reduced expression while those flanking the ETS element had no effect or minimal effect. (E) ETS1 activation of ATXN2 depends on the ETS-binding site. Overexpression of ETS1 increased ATXN2-luc. Inclusion of the D14 deletion in ATXN2-luc blocked the ability for ETS1 to increase ATXN2-luc expression. Values are means and SD from three independent transfections each read in triplicate and expression was controlled with TK-Renilla luciferase.