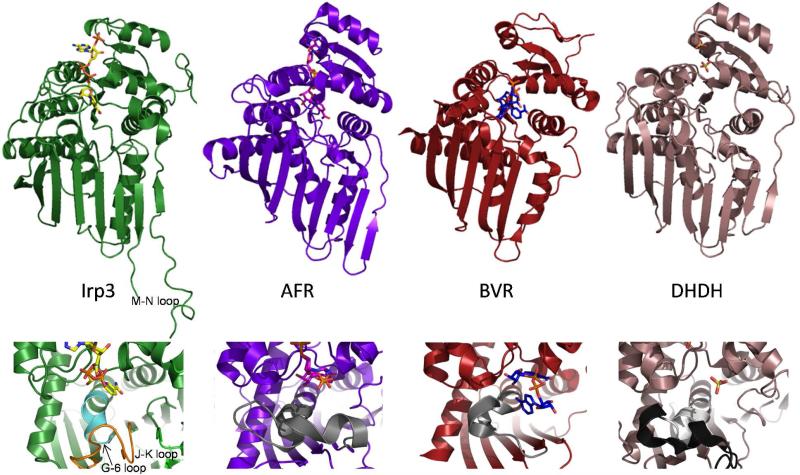
Figure 4. Cartoon representations of the NADP+-bound Irp3, 1,5-anhydro-D-fructoase reductase (AFR, Protein Data Bank code 2GLX), biliverdin reductase (BVR, code 1LC3), dimeric dihyrodiol dehydrogenase (DHDH, code 2O48).
The upper panels show the overall fold in cartoon and the NADP+ (or sulfates in DHDH) in sticks. It should be noted that the NADP+ binds in a similarly extended conformation in Irp3 and AFR. In BVR, the NADP+ is folded in a U-shaped conformation that is hypothesized to be the result of crystal packing (35). The DHDH structure does not contain NADP+, but has sulfates bound in locations similar to where the phosphates of NADP+ are anticipated to bind (30). In Irp3, the M-N loop, which has 6 to 15 residues disordered depending on monomer, is labeled. The lower panels are a close-up of the putative substrate binding region. In DHDH, the region of the GGX3DX3(Y/H) consensus sequence is shown in light grey and the remainder of the loop containing this sequence is black. The corresponding loops in AFR and BVR are also grey. In Irp3, the much shorter loop connecting strand G and helix 6 is shown in cyan. The extended loop connecting the J and K strands that occupies the corresponding space in three-dimensions is shown in orange.