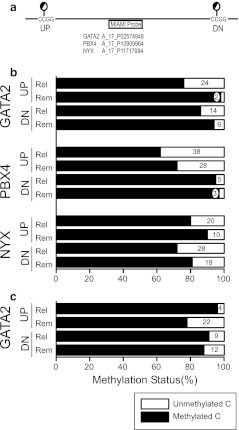
Fig. 3.
DNA methylation ratio analysis by bisulfite-pyrosequencing. To validate the MIAMI data, methylation ratios for CpGs in the closest CCGG HpaII recognition sites on both the 5′ (UP) and 3′ (DN) side of the probe sequences were determined using bisulfite-pyrosequencing (a). These ratios were lower in minimal change nephrotic syndrome (MCNS) relapse (Rel) samples than in remission (Rem) samples at all sites for the three probes in Th0s (b). The methylation ratios for the GATA2 probe A_17_P02574948 in monocytes (c) were higher in Rel than in Rem at both sides of the probe. The methylation pattern at every CpG site determined using bisulfite-pyrosequencing accorded well with the MIAMI results (Table 3), which indicate an unmethylated intensity ratio between relapse and remission