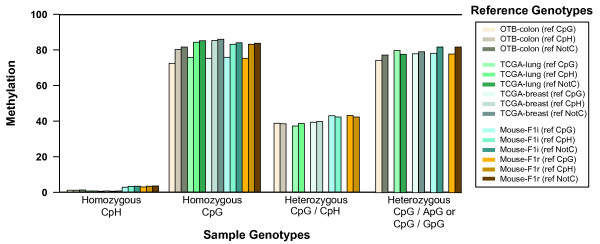
Figure 5.
Accurate methylation calling at SNPs. Bis-SNP was run on five different datasets, single-end sequencing from Colon Mucosa Tissue [6] (a), two TCGA samples using paired-end sequencing from breast and lung tissues (normal, non-cancer), and two mouse samples using paired-end sequencing from [22] (see Table 1). In each case, Bis-SNP was used to identify cytosines in one of four sequence context in the sample genome. For each sample genotype, cytosines were further divided by their sequence context in the reference genome ('ref CpG', 'ref CpH', or 'refNotC'). All cytosines within a particular category in a particular sample were averaged to yield a mean methylation level. The number of cytosines in each category can be found in Table 2.