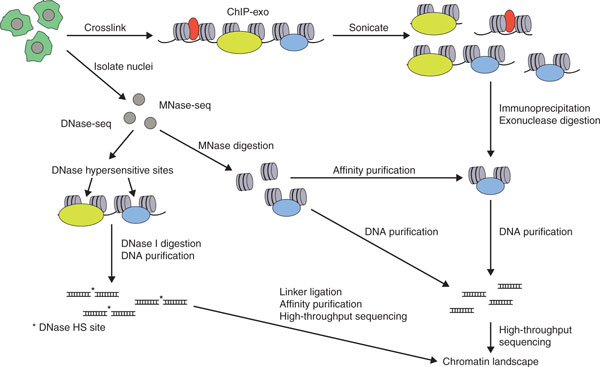
Figure 2.
Summary of techniques for base-pair resolution epigenome mapping. Schematic representations of ChIP-exo, MNase-seq, and DNase-seq. In ChIP-exo, chromatin is sonicated and specific fragments are isolated with an antibody to a protein of interest. ChIP DNA is trimmed using λ exonuclease, purified, and sequenced. In MNase-seq, nuclei are isolated and treated with MNase to fragment chromatin. Chromatin is then subjected to DNA purification with or without prior affinity purification and MNase-protected DNA is sequenced. In DNase-seq, nuclei are isolated and treated with DNase I to digest chromatin. DNase-hypersensitive DNA is then ligated to linkers, affinity purified, and sequenced. HS, hypersensitive.