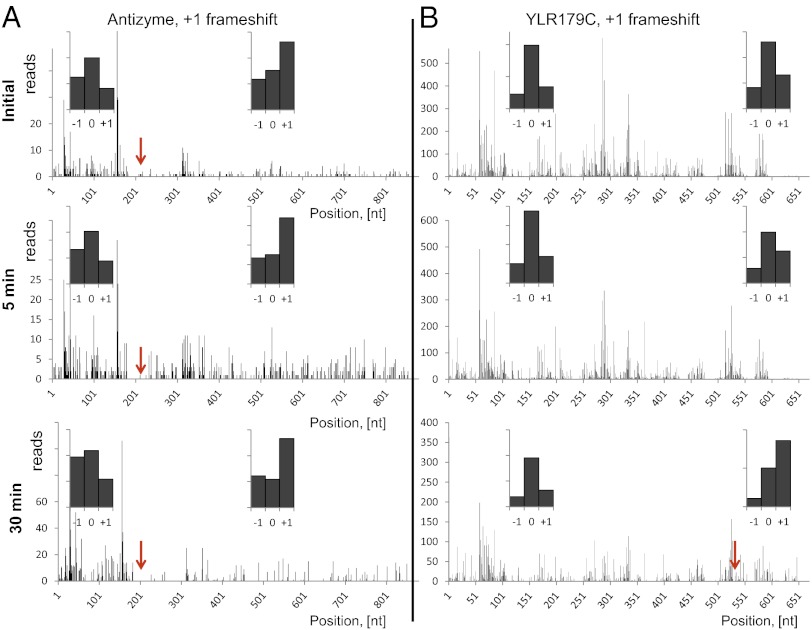
Fig. 3.
Ribo-seq allows identification of frameshifts (red arrows). (A) Validation of the known frameshift in the antizyme gene. (B) Oxidative stress leads to a frameshift in the product of the YRL179C gene. We observed a change of frame, leading to translation of a longer protein in the 30-min peroxide treatment sample. The 5′ ends of footprints were mapped to the genomic sequence of YRL179C. (Insets) Histograms show the count of footprints, matching one of three possible frames either to the left or to the right of the frameshift. The “0” frame is the one with the annotated start codon. The highest count of footprints matched the “0” frame before the frameshift and the “+1” frame after the frameshift.