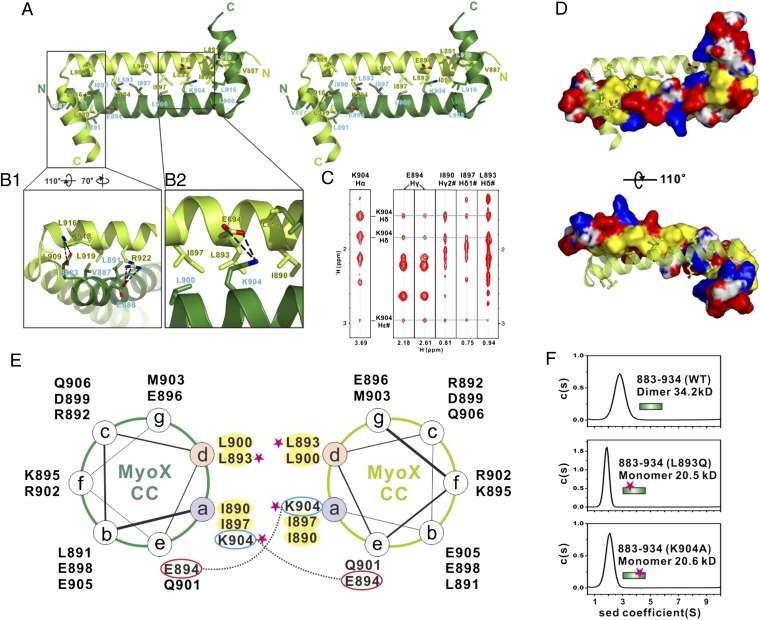
Fig. 3.
Molecular details of the Myo10 anti-CC dimer. (A) Stereoview showing the dimerization interface of the Myo10 anti-CC dimer. The side chains of the residues involved in the dimer interface are drawn in the stick model. (B) Detailed interactions showing the interface of αB with the N-terminal end of αA (B1) and the interface of K904 with residues from the neighboring subunit (B2). (C) Selected strips from the 3D 13C-NOESY spectrum of Myo10 (aa 883–934) showing long-distance NOEs between the side chain of K904 with the side chains of E894, I890, I897, and L893. (D) Combined-surface and ribbon representation of the Myo10 anti-CC dimer. In the surface diagram, the positively charged amino acids are drawn in blue, the negatively charged residues in red, the hydrophobic residues in yellow, and the others in white. (E) Helical wheel presentation showing the heptad repeats at the center of αA. Interhelical salt bridges between the residues at a and e positions are depicted by dashed lines. Residues at “a” and “d” positions forming the hydrophobic core of the coiled coil are highlighted in yellow. Single substitution mutations that disrupt the Myo10 dimer formation are highlighted with pink stars. (F) Sedimentation velocity analysis showing that the L893Q and the K904A mutants of anti-CC are monomer in solution.