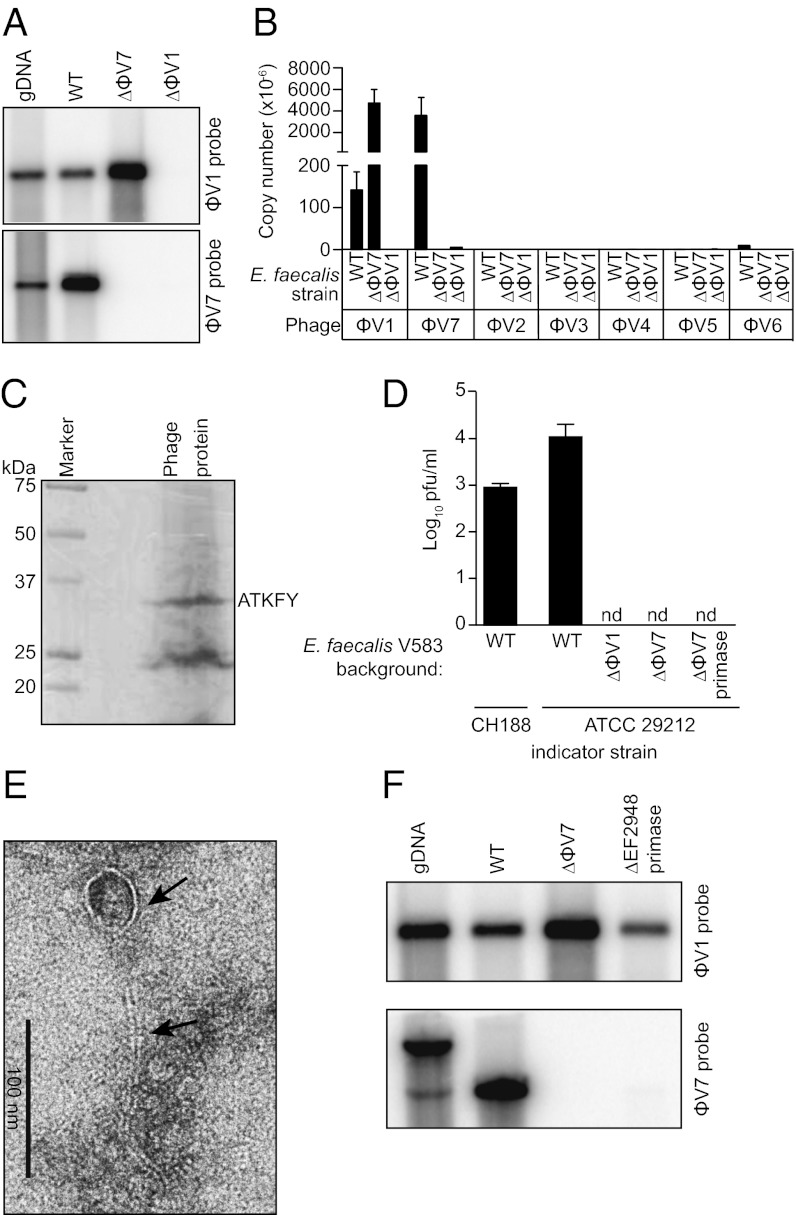
Fig. 2.
E. faecalis V583 produces a ϕV1/7 composite bacteriophage. (A) Southern blot of phage DNA isolated from the culture supernatant of WT E. faecalis V583 (WT) or the isogenic mutant strains ΔϕV7 and ΔEF0339 (ΔϕV1 capsid protein). DNA was digested with NdeI (ϕV1 probe) or NsiI (ϕV7 probe). WT V583 genomic DNA (gDNA) is used as a control. (B) Quantification of phage DNA isolated from WT or ϕV1/7-deficient E. faecalis culture supernatant. The absolute copy number of each prophage was measured using qPCR. (C) Imido black staining of ϕV1/7 phage proteins separated by SDS/PAGE. N-terminal sequencing identified a ∼32 kDa protein as the EF0339 ϕV1 capsid protein. (D) Quantitative plaque assay measuring ϕV1/7 particles from E. faecalis V583 or the ΔϕV1, ΔϕV7, and ΔϕV7 primase mutants using E. faecalis CH188 or ATCC 29212 as indicator strains. nd, not detected. (E) Transmission electron micrograph of a ϕV1/7 particle. Arrows indicate the phage capsid and tail. (F) Southern blot of phage DNA isolated from the culture supernatant of the ϕV7 ΔEF2948 primase mutant E. faecalis strain. WT V583 genomic DNA and phage DNA from WT V583 and ΔϕV7 mutant cultures were probed for comparison. The genomic DNA band detected by the ϕV7 probe has a higher molecular weight because of a chromosomal NsiI site that is outside of the ϕV7 genome.