Fig 4.
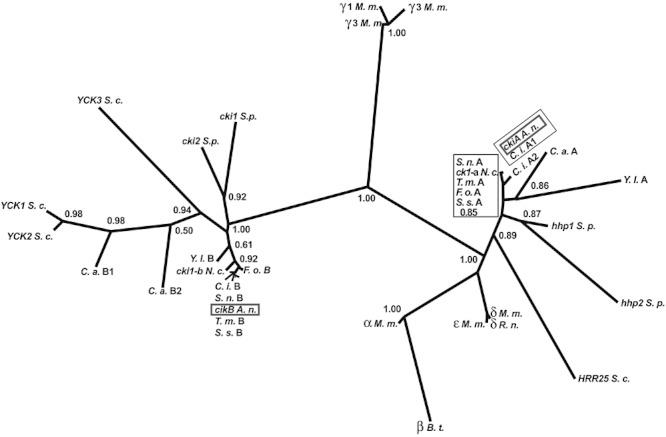
Phylogeny of the ascomycete casein kinase I proteins. This tree includes casein kinases I from representative species of the ascomycetes. All the isoforms of Mus musculus (M. m.) are also included, together with the unique isoform β of Bos taurus (B. t.) and the isoform δ of Rattus norvegicus (R. n.) mentioned in various contexts in the text. Characterized genes of A. nidulans, N. crassa, S. cerevisiæ and S. pombe are indicated by their standard genetic symbols followed by the abbreviation of the species. Other ORFs are indicated by the following convention: A two letter abbreviation of the species followed by ‘A’ for the homologues of ckiA, HRR25, hhp1 and hhp2, which never have a palmitoylation signal in the their carboxy-terminus, and ‘B’ for the homologues of YCK which always carry such a signal. When more than one ‘A’ or ‘B’ homologue is present for a given species, this indicated by a number following the letters A or B. The ascomycete species included are: Taphrinomycotina: Schizosaccharomyces pombe (S. p.); Saccharomycotina, Saccharomycetales: Saccharomyces cerevisiæ (S. c.), Candida albicans (C. a.), Yarrowia lypolitica (Y. l.). Pezizomycotina: Eurotiales, Aspergillus nidulans (A. n.), Onygenales, Coccidioides immitis (C. i.), Sordariales, Neurospora crassa (N. c.), Hypocreales, Fusarium oxysporum (F. o.), Helotiales, Sclerotina sclerotium (S. s.), Pleosporales, Stagonospora nodorum (S. n.), Pezizales, Tuber melansporum (T. m.). ORF sequences were obtained from the appropriate databases (see Supplementary Experimental procedures for accession numbers) and aligned with T-Coffee. The phylogenetic tree was redrawn from an original PhyMyl (Guindon et al., 2010) tree obtained after curation of the alignment with G-blocks and visualized with Drawtree (see Experimental procedures). The decimals in the nodes indicate the values of the Approximate Probability Ratio Test (aLTR; Anisimova and Gascuel, 2006). For clarity's sake some of the values for the minor nodes are not included. The boxed homologues are not separated in this tree, the 0.85 value refers to the aLTR separating the two boxed branches containing the Pezizomycotina homologues in clade ‘A’. The non-boxed homologues in a column in the ‘B’ clade are separated by extremely short branches, they correspond to up to bottom to the five short branches, left to right. Note the very high branch support for all the major nodes. The two A. nidulans genes are highlighted by a grey box. Note the striking conservation of sequences within the Pezizomycotina, including Tuber melanosporum, a basal species (Spatafora et al., 2006; see Fig. 4), which has diverged from the crown orders of the Pezizomycotina (including all the other Pezizomycotina represented in the figure) about 300 million years ago (Berbee and Taylor, 2007; Lucking et al., 2009).
