Figure 1.
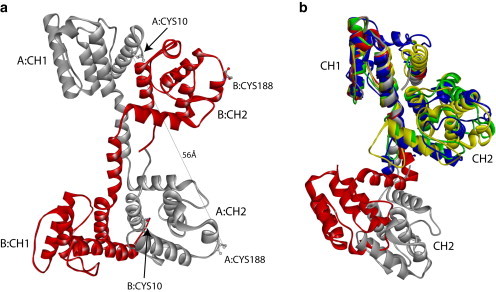
(a) X-ray crystal structure of the N-ABD of human dystrophin (1DXX.pdb) indicates an antiparallel, domain-swapped dimer (5). The two monomers, labeled A and B, are shown in gray and red, respectively. In the dimer, the CH1 domain from one monomer interacts with the CH2 domain from the other monomer. Each monomer contains two cysteines (Cys-10 and Cys-188), one in each CH domain, at an intramolecular distance of 56 Å. (b) Structural alignment of x-ray crystal structures of tandem CH domains of dystrophin (1DXX.pdb; gray), utrophin (1QAG.pdb; red), fimbrin (1AOA.pdb; blue), α-actinin (1TJT.pdb; green), and plectin (1MB8.pdb; yellow) using the MultiProt program (47) (http://bioinfo3d.cs.tau.ac.il/MultiProt/). Dystrophin and utrophin tandem CH domains crystallize in an open conformation where the two CH domains do not interact, whereas tandem CH domains of fimbrin, α-actinin, and plectin crystallize in a closed conformation with significant inter-CH domain interactions. The N-terminal (CH1) and C-terminal (CH2) CH domains are labeled in the figure. Molecular structures were drawn using the program Accelrys Discovery Studio Visualizer (http://accelrys.com/products/discovery-studio/visualization-download.php).
