Figure 3.
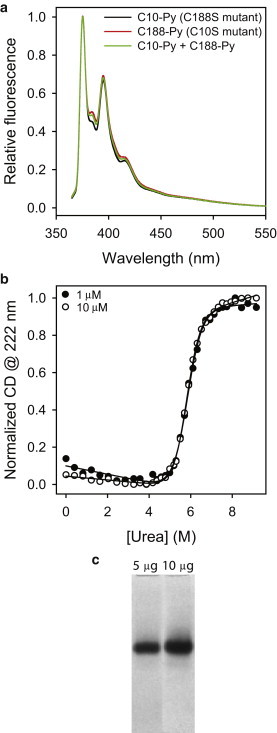
Dystrophin N-ABD is a monomer in solution. (a) Pyrene fluorescence of single-cysteine mutants (C188S (C10-Py) and C10S (C188-Py); 10 μM), and when mixed at equimolar ratio (C10-Py + C188-Py; 10 μM total protein concentration). The absence of excimer fluorescence indicates the absence of dimers or higher oligomers in solution. (b) Thermodynamic stability of dystrophin N-ABD measured at 1 and 10 μM. Urea was used to unfold the protein, and the CD at 222 nm was used to monitor unfolding of the α-helical secondary structure of the protein. The stability is independent of the protein concentration confirming the absence of stable dimers at or below 10 μM protein concentration. (c) Native PAGE of dystrophin N-ABD with 5 and 10 μg loading protein amounts. A similar result was observed even when the gel was overloaded with 30 μg of protein (7).
