Abstract
Background.Biomarkers of progression from latent Mycobacterium tuberculosis infection to active tuberculosis are needed. We assessed correlations between infection outcome and antibody responses in macaques and humans by high-throughput, proteome-scale serological studies.
Methods.Mycobacterium tuberculosis proteome microarrays were probed with serial sera from macaques representing various infection outcomes and with single-point human sera from tuberculosis suspects. Fluorescence intensity data were analyzed by calculating Z scores and associated P values. Temporal changes in macaque antibody responses were analyzed by polynomial regression. Correlations between human responses and sputum bacillary burden were assessed by quantile and hurdle regression.
Results.Macaque outcome groups exhibited distinct antibody profiles: early, transient responses in latent infection and stable antibody increase in active and reactivation disease. In humans, antibody levels and reactive protein numbers increased with bacillary burden. Responses to a subset of 10 proteins were more tightly associated with disease state than reactivity to the broader reactive proteome.
Conclusions.Integration of macaque and human data reveals dynamic properties of antibody responses in relation to outcome and leads to actionable findings for translational research. These include the potential of antibody responses to detect acute infection and preclinical tuberculosis and to identify serodiagnostic proteins for the spectrum of bacillary burden in tuberculosis.
The vast majority of persons infected with Mycobacterium tuberculosis are asymptomatic. In these individuals, who are estimated to constitute one-third of the human population, tubercle bacilli presumably persist in very low numbers. Only >10% of immunocompetent, latently infected individuals develop active disease and become infectious. Nevertheless, M. tuberculosis infection causes 9.4 million new cases of active tuberculosis and 1.7 million deaths per year [1]. Transmission of infection would be greatly reduced if it were possible to identify and treat infected individuals as they progress to active disease before they become symptomatic and infectious. Detecting tubercle bacilli or bacillary products is exceedingly difficult during preclinical disease due to low bacillary numbers. Thus, we tested the hypothesis that host-derived biomarkers track the course of infection with M. tuberculosis.
We focused on the antibody response as a potential biomarker of progression of M. tuberculosis infection for several reasons. First, serum levels of specific antibodies are typically detected during active tuberculosis but not during stable latent infection [2], indicating stage-specific responses. Second, we observed changes in the protein composition of the seroreactive proteome of M. tuberculosis associated with disease [3], indicative of correlations between bacillary antigen production and antibody targets. Third, temporal changes of the antibody response prior to clinical manifestation and active tuberculosis diagnosis have been observed in human immunodeficiency virus (HIV)–coinfected individuals [4, 5]. For the reasons above, it should be possible to find serological markers of infection outcome and tuberculosis reactivation.
To examine global changes in the antibody response associated with infection outcome and disease progression, we utilized high-throughput proteome microarray technology [6] and 2 host systems: experimental infection of macaques and human tuberculosis. The macaque model is relevant to human tuberculosis because it well recapitulates the various outcomes of M. tuberculosis infection seen in humans, including spontaneous reactivation [7, 8]. Thus, the pathogenesis of macaque and human tuberculosis is similar, even though immunological differences between macaques and humans exist (eg, [9–11]) and comparative studies of immune responses to tuberculosis in the 2 species are still lacking. Further, parallel studies are warranted because these 2 host systems complement each other. On one hand, it is possible to monitor temporal changes of the antibody response to M. tuberculosis infection with the macaque model, whereas conducting longitudinal human studies is exceedingly difficult, even in high-burden countries, due to the low frequency of reactivation in immunocompetent individuals. On the other hand, correlations between antibody levels and bacillary burden (an indicator of disease severity) are best assessed in humans. Enumeration of acid-fast bacilli in sputum of tuberculosis suspects is common clinical practice and is a reasonable surrogate of lung bacillary burden. In contrast, bacterial counts are not routinely performed in macaques because enumerating tubercle bacilli requires sacrifice of these expensive animals.
Here, we analyzed proteome-scale antibody responses in serial sera from infected macaques representing different infection outcomes and in sera from tuberculosis patients and controls in relation to sputum bacillary burden. We report the parallel characterization of the macaque and human antibody response to M. tuberculosis infection at the proteome scale. Moreover, by integrating monkey and human global measurements, we find that the antibody response changes quantitatively and qualitatively with M. tuberculosis infection outcome and disease severity in both hosts.
MATERIALS AND METHODS
Experimental Animals
Sera from 14 cynomolgus macaques of Philippine or Chinese origin were used. These macaques exhibit greater immunogenetic diversity than those from other geographic regions, such as the Mauritian cynomolgus [9, 12, 13]. Protocols for infection and clinical and bacteriological assessments were as published [7, 14]. Three criteria of outcome-based grouping were used: (1) active disease: persistent evidence of disease, with ongoing radiographic involvement, persistent culture positivity, or other clinical signs of active disease; (2) latent infection: no radiographic involvement after 4 weeks of infection and no clinical sign of disease for the study period (1–3 years); and (3) reactivation disease: similar to latent disease but developed active disease spontaneously following an initial, disease-free period lasting at least 6 months. In the present work, spontaneous reactivation was caused by transfer between housing facilities.
Human Sera
Sera utilized to probe proteome microarrays were from a retrospective serum bank collected from adults in 2003–2008 in the context of a National Institutes of Health–funded, international, multisite study titled “Clinical Suspicion of Tuberculosis” (PI: A. Catanzaro, acatanzaro@ucsd.edu). Recruitment of tuberculosis suspects was based on epidemiologic factors, symptoms, and radiographic findings under uniform protocols approved by institutional ethics committees at each site. Final diagnosis of active tuberculosis was based on positive M. tuberculosis culture. Sputum-smear status of active tuberculosis patients was based on Ziehl–Neelsen staining results. Diagnosis of nontuberculosis disease (NTBD) was based on negative M. tuberculosis culture plus a positive diagnosis for other disease. Seven percent of all study subjects were positive for HIV infection. Here we utilized sera from 397 tuberculosis suspects, 169 of whom were diagnosed with active tuberculosis (tuberculosis patients) and 228 of whom received an alternative diagnosis (NTBD patients). Mean age of tuberculosis and NTBD patients were 49 (±17) and 40 (±17), respectively. The countries of serum collection were Philippines (45%), the United Kingdom (27%), Mexico (20%), and the United States (8%). Previous history of active tuberculosis was reported for 31% of NTBD and 17% of tuberculosis patients. Sera were collected within 1 week of antituberculosis chemotherapy, when applicable.
Proteome Microarrays
Mycobacterium tuberculosis proteome microarrays were manufactured and probed with sera following published protocols [3]. On the arrays, serum reactivity to each protein is detected as intensity of the fluorescence signal. Analytical performance characteristics and reproducibility of the assay have been reported [3]. Limitations of the system, such as lack of posttranslational modifications of proteins expressed in recombinant Escherichia coli, have been discussed [3]. Study protocols were approved by the institutional review board at the University of Medicine and Dentistry of New Jersey.
Assessment of Serum Reactivity
Given the proteome array characteristics (eg, different amount of protein per spot), each protein spot was treated as an independent assay, and Z statistics were used to identify protein spots that reacted with serum, as previously described [3]. Null distributions for the Z statistics were obtained from preinfection sera (n = 14) in the macaque study and from sera from noninfected individuals (defined by negative tuberculin skin test) (n = 14) in the human study. A P value ≤ .01 after correction for multiple testing was used to define a serum as reactive to a protein (reactivity call).
Enrichment Analysis
Fisher exact tests were performed with 2 sets of protein classifications: subcellular localization by LocateP (http://www.cmbi.ru.nl/locatep-db/cgi-bin/locatepdb.py) and functional classification by TubercuList (http://tuberculist.epfl.ch/).
Modeling of Antibody Responses in Macaques
Locally estimated scatterpoint smoothing (LOESS) regression fits a model to a scatter plot by estimating polynomial regression models for each data point using a small proportion of nearby (local) data points [15]. Polynomial regression models of antibody responses over time were estimated up to 650 days postinfection. The time terms included in the models were suggested by the fluctuations of the time trajectory in the LOESS regression. To compensate for the lack of independence among successive antibody measurements over time, we used generalized estimating equations, assuming an autoregressive correlation structure [16]. To reduce the intrinsic correlation between powers of time, these powers were transformed to orthogonal polynomials.
Modeling of Antibody Responses in Tuberculosis Suspects
Two regression modeling approaches were used with the human data. Quantile regression, which models the complete distribution, including median and extremities [17], was used because serum reactivity is seen at the high tail of the distribution in the microarray platform [3]. Hurdle models include a logistic component modeling the probability of a response and a truncated Poisson component modeling response magnitude [18].
Analytical Software
R (http://www.r-project.org) was used for reactivity call estimation and quantile regression analysis; TMEV (http://www.tm4.org) was used for enrichment analysis; and SAS (http://www.sas.com/) was used for polynomial and hurdle regression models.
RESULTS
Reactivity of Macaque Sera With M. tuberculosis Proteins
To investigate antibody responses during infection, we probed M. tuberculosis proteome microarrays with serial sera collected from experimentally infected macaques with active disease, latent infection, and reactivation disease (4 or 5 animals per infection outcome group). For each animal, we tested serum from 1 preinfection time point and from approximately 10 postinfection time points at 1-month intervals. Using data collected from preinfection sera as the null distribution, we defined 101 proteins as reactive to at least 1 postinfection serum, with a false discovery rate ≤1% (Supplementary Table 1). The reactive protein pool defines the immunoproteome. Protein class analysis showed that the immunoproteome was enriched for extracellular proteins and Pro-Glu/Pro-Pro_Glu (PE/PPE) family proteins [19] (Fisher exact test, P < .01), in agreement with previous observations that these protein classes are frequent immunological targets [20].
Having established size and composition of the immunoproteome, we next examined serum reactivity per animal. We observed that, even though the animals had been infected under the same protocol, antigen recognition varied considerably from 1 animal to another (Figure 1A). Thus, the observed variation in antibody responses is host driven. Results in Figure 1A also indicated that sera from the active and the reactivation groups reacted with more proteins than those from the latent group (22, 10, and 5 proteins on average, respectively) (Supplementary Table 1), suggesting that infection outcome was reflected in the antibody response. This association is examined below.
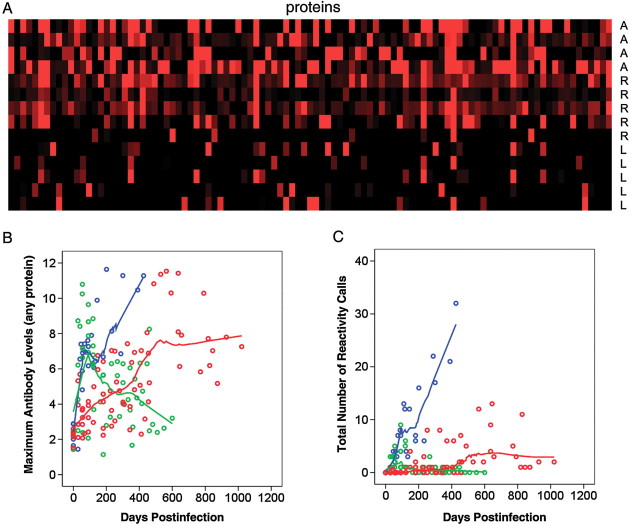
Figure 1.
Antibody responses in cynomolgus macaques. Fourteen cynomolgus macaques were infected intratracheally with approximately 25 colony-forming units of Mycobacterium tuberculosis strain Erdman per animal. Preinfection sera and approximately 10 postinfection sera per animal were tested with proteome microarrays. The number of sera per macaque varied depending on the duration of follow-up, which was shortest for the animals in the active class. Based on Z statistics, 101 proteins were identified as significantly reactive to postinfection sera, compared with pre-infection sera. A, Maximal antibody levels obtained at any time point to the 101 reactive proteins (columns) are shown. The data were normalized to preinfection sera signal intensities by Z score estimation; the red color indicates Z score = 2, and the black color indicates Z score = 0. A, active class; L, latent class; R, reactivation class. B, Maximal antibody levels over time. Each circle represents maximal antibody level to any of the 101 reactive proteins in a particular macaque. Outcome groups are color-coded: blue, active; green, latent; red, reactivation. Locally estimated scatterpoint smoothing (LOESS) regression curves are shown. C, Numbers of reactive proteins over time. Each circle represents the total number of reactive proteins in a particular macaque. As in B, outcome groups are color-coded, and LOESS regression curves are shown.
Temporal Changes in Antibody Responses in Relation to Infection Outcome
Two parameters of the antibody response were analyzed: antibody levels and number of reactive proteins. Given the heterogeneity of antigen recognition, we used maximal antibody levels against any 1 of the 101 reactive proteins. Maximal antibody levels were plotted against time after infection and stratified by outcome class, and LOESS curves were fitted to these plots (Figure 1B). In the latent infection group, antibody levels were characterized by an early, transient rise followed by a decline to preinfection levels. In contrast, in the active disease class, the early rise was followed by a further increase of antibody levels. A slower increase characterized the response in the reactivation class. Similar changes were observed when antibody responses were expressed as number of reactive proteins (Figure 1C).
To determine whether the temporal changes of the antibody response depicted by the LOESS curves were statistically significant, we estimated polynomial regression models. We first analyzed maximal antibody levels. Because the LOESS curves for active disease and reactivation classes included 1 local maximum and 1 local minimum, which correspond to a third-degree polynomial, we modeled antibody responses in these 2 classes by including the terms Time, Time2, and Time3. As shown in Table 1, the statistically significant coefficients of Time indicated that antibody levels in both classes increased over time and that the increase was more rapid with active tuberculosis than with reactivation (7.88 vs 1.91). The statistically significant coefficients of Time, Time2 and Time3 for the active tuberculosis group, together with a positive value of the discriminant of the polynomial's derivative (ie, 3.182–3 × 7.88 × 0.41 > 0), imply an initially increasing antibody response that may transiently weaken (local maximum, followed by local minimum) but then resumes an overall increasing trend. In contrast, the antibody response in the reactivation group showed only a monotonic increase because Time2 and Time3 coefficients were nonsignificant. The response in the latent infection class was modeled separately because the LOESS curve showed 2 local maxima, which suggested a quartic time term (Time4). Models estimated for this class showed that overall antibody levels initially increased over time (Table 1; Time effect), subsequently fell, rose again, and finally declined. All these fluctuations were statistically significant (Table 1; Time2, Time3, and Time4 effects). The models estimated for maximal antibody levels also applied to changes in the number of reactive proteins over time (Table 1), suggesting that these 2 properties of the antibody response changed concurrently. Collectively, the results show that changes of the antibody response as a function of time after infection were associated with infection outcome.
Table 1.
Polynomial Regression Models of Antibody Responses vs Time Since Infection
| Parameter | Reactivation | Active | Latent |
|---|---|---|---|
| Maximal antibody levelsa as outcome variable | |||
| Time | 1.91 (.35–3.47) | 7.88 (5.08–10.67) | 10.58 (6.38–14.78) |
| Time2 | −0.36 (−1.00 to .28) | −3.18 (−4.99 to −1.37) | −7.69 (−10.91 to −4.46) |
| Time3 | 0.033 (−.04 to .11) | 0.41 (.11 –.72) | 1.89 (1.02–2.76) |
| Time4 | NA | NA | −0.15 (−.22 to −.07) |
| Number of reactive proteinsb as outcome variable | |||
| Time | 1.21 (.12–2.29) | 6.53 (4.38–8.69) | 5.28 (2.21–8.35) |
| Time2 | −0.28 (−.73 to .16) | −2.71 (−4.19 to −1.24) | −3.83 (−6.20 to −1.46) |
| Time3 | 0.03 (−.02 to .08) | 0.36 (.10 –.61) | 0.95 (.30–1.59) |
| Time4 | NA | NA | −0.08 (−.13 to −.02) |
Polynomial regression analyses were performed with maximal antibody levels or number of reactive proteins as outcome variable and Time (linear), Time2 (quadratic), Time3 (cubic), or Time4 (quartic) as independent variables. The estimated regression coefficients and 95% confidence intervals are shown. Confidence intervals that exclude the value zero indicate a statistically significant regression coefficient (P < .05; shown in bold). Models that include a positive, statistically significant linear coefficient define a curve that is increasing over time; the other coefficients (quadratic, cubic and quartic terms) represent undulations in the curve. If these latter coefficients are not statistically significant then the departures from a straight-line model (ie, the undulations) are due to chance (ie, random error), implying that the observed data is consonant with a strictly increasing linear function of time. Transformation of powers of Time to orthogonal polynomials, which was tested to reduce the intrinsic correlation between powers of Time, estimated models extremely similar to those based on untransformed powers of Time (not shown).
Abbreviation: NA, not applicable (not estimated).
a Maximal antibody levels, the highest normalized signal intensity obtained with any of the 101 reactive proteins.
b Log transformed.
Antibody Responses in Active Tuberculosis Patients
Because antibody levels can be sensitive markers of antigen burden [21], we reasoned that the association between antibody responses and infection outcome in macaques could reflect the effect of bacillary burden on antibody responses. We tested this association in human tuberculosis by probing proteome microarrays with sera collected from patients with active tuberculosis (n = 169) and NTBD (n = 228).
We first identified proteins reactive with human sera. By applying Z statistics and a false discovery rate of ≤0.01 (see Methods), we identified 356 proteins reactive to at least 1 tuberculosis-suspect serum (Supplementary Table 2). This immunoproteome was rich in extracellular proteins, proteins of cell wall and cell processes, PE/PPE proteins, and proteins involved in lipid metabolism (Fisher exact test, P < .01). Almost 30% of the macaque immunoproteome was contained in the human counterpart (Supplementary Tables 1 and 2). Within the human immunoproteome, reactivity to 10 proteins was significantly associated with active tuberculosis (tuberculosis-associated proteins) (Table 2). Most (8 of 10) tuberculosis-associated proteins identified in the present study were also identified as tuberculosis associated in an earlier, independent multisite study [3], strongly indicating the existence of universal immunodominant antibody targets.
Table 2.
Proteins Associated With Active Tuberculosis
| Protein ID | Odds Ratio (95% CIa) | Annotationsb |
|---|---|---|
| Rv0934c | 7 (2.2–28.7) | Glycolipoprotein 38 kDa antigen/PstS1 |
| Rv1174c | 9.6 (1.2–434.5) | Low molecular weight T-cell antigen/TB8.4 |
| Rv1411cc | 7.7 (2.1–41.8) | Lipoprotein LprG |
| Rv1860c | 15.5 (2.2–671.4) | Secreted glycoprotein 45–47 kDa antigen/MPT32 |
| Rv1980cc | 9.6 (1.2–434.5) | Secreted antigen MPT64 |
| Rv3616cc | 11 (1.4–492) | Conserved hypothetical protein |
| Rv3763 | 12.5 (1.7–552.2) | Lipoprotein lpqH/19 kDa Antigen |
| Rv3804cc | 5.4 (2.1–16.8) | Secreted antigen 85A/mycolyltransferase |
| Rv3874c | 12.4 (2.9–111.9) | Secreted antigen Cfp10/EsxB |
| Rv3881cc | 4.6 (1.7–14.4) | SecretedEspB |
All proteins were previously identified as B-cell or T-cell antigens.
Abbreviation: CI, confidence interval.
a Odds ratios were calculated from reactivity calls.
b Annotations were adapted from the Sanger Institute database (http://www.sanger.ac.uk/Projects/M_tuberculosis/Gene_list/).
c They have been already identified as active tuberculosis associated in our previous proteome-scale serological screening [3].
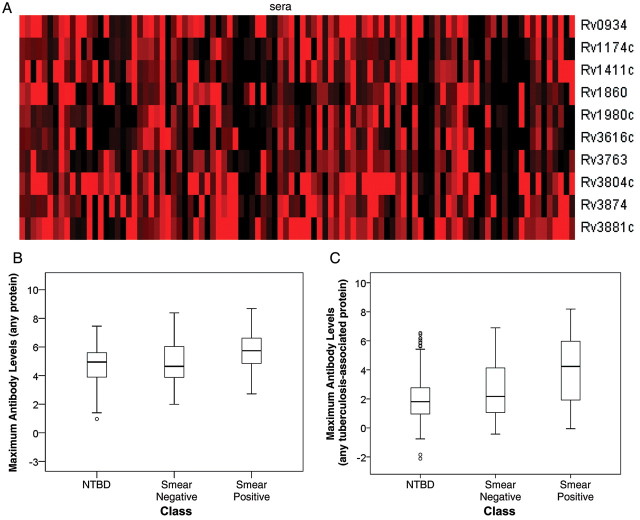
We next asked how antibody levels correlated with bacillary burden. For this analysis, we stratified tuberculosis suspects into 3 groups of no (NTBD), low (smear-negative tuberculosis), and high (smear-positive tuberculosis) burden. As in previous work with human sera [3] and with the macaque data (Figure 1A), antigen recognition varied from 1 patient to another (Figure 2A). Thus, we used maximal antibody levels to the immunoproteome to summarize antibody response per subject. We observed higher antibody levels in the low- and high-burden classes than in the no-burden class (Figure 2B). Differences among groups were even higher when maximal antibody levels to only the 10 tuberculosis-associated proteins were analyzed (Figure 2C). We next investigated the statistical significance of the observed differences by quantile regression analysis. We found that the 60th, 70th, and 80th quantiles consistently showed that maximal antibody levels in the smear-positive class were significantly higher than in the NTBD class after controlling for age, country of origin, and past tuberculosis (Table 3). When only maximal levels of antibody to the tuberculosis-associated proteins were analyzed, significantly higher antibody levels relative to the no-burden class were observed for both low- and high-burden classes (Table 3).
Figure 2.
Antibody levels in tuberculosis suspects. The Mycobacterium tuberculosis proteome arrays were probed with sera from 169 active tuberculosis and 228 nontuberculosis disease (NTBD) patients. Reactive proteins (n = 356) were identified by Z statistics, and proteins associated with active tuberculosis (n = 10) were identified by odds ratio calculation. A, Heat map of reactivity (Z scores) to 10 tuberculosis-associated proteins (rows) is shown for reactive sera from tuberculosis patients (columns); the red color indicates Z score = 3 and the black color indicates Z score = 0. B, Distribution of maximal antibody levels to any of the 356 reactive proteins by diagnostic class. The box plots show the median of the distribution (horizontal line in the box), the interquartile range (box), the range excluding outliers (whiskers), and the outliers (circles). C, Distribution of maximal antibody levels to any of the 10 tuberculosis-associated proteins by diagnostic class. Data are presented as box plots, as in B.
Table 3.
Quantile Regression Models of Maximal Antibody Levels vs Bacillary Burden
| Regression Coefficients |
|||
|---|---|---|---|
| Bacillary Burden | 60th Quantile | 70th Quantile | 80th Quantile |
| Maximum antibody levels to any reactive protein | |||
| None (reference) | |||
| Low | −0.19 (−.49 to .61) | 0.08 (−.40 to .82) | 0.53 (−.08 to .89) |
| High | 0.97 (.62–1.31) | 0.97 (.69–1.25) | 0.78 (.48–1.12) |
| Maximum antibody levels to any tuberculosis-associated protein | |||
| None (reference) | |||
| Low | 0.66 (−.08 to 1.30) | 0.83 (.31–1.63) | 1.08 (.53–2.25) |
| High | 2.47 (2.10–3.14) | 2.74 (2.41–3.18) | 2.61 (2.29–3.19) |
Quantile regression was performed with maximum antibody levels to any reactive protein or tuberculosis-associated protein as dependent variable and burden class as independent variable. Regression coefficients (and 95% confidence intervals in parentheses) were calculated for the 60th, 70th and 80th quantiles after adjusting for age, country of serum sampling, and history of past tuberculosis. Bacillary burden: none, nontuberculosis disease; low, sputum smear-negative tuberculosis; high, sputum smear-positive tuberculosis. The coefficients indicate antibody levels in either burden class compared with the no-burden class (reference). Differences between burden classes are significant (shown in bold) when the confidence intervals of coefficients do not include zero and when confidence intervals of 2 burden classes do not overlap.
The proteome-wide platform also gave the opportunity to assess the number of reactive proteins as another parameter of the antibody response, as tested in macaques. We used hurdle regression to model the probability to react to at least 1 protein and the number of antigenic targets in reactive sera in relation to sputum bacillary burden. When we analyzed reactivity to the immunoproteome, we found that the odds of a response to at least 1 protein were 3-fold (95% confidence interval [CI], 1.79–4.81) higher in subjects with smear-positive tuberculosis than in those with NTBD, whereas no difference was seen between smear-negative tuberculosis and NTBD (data not shown). Moreover, the mean number of protein targets in seroreactive tuberculosis patients, regardless of smear status, was only 1.5-fold (95% CI, 1.32–1.87) higher than that in NTBD subjects. In contrast, when we analyzed reactivity to the tuberculosis-associated proteins, the odds of a response were 12-fold (95% CI, 6.07–22.21) higher in smear-positive tuberculosis than in NTBD subjects and 3-fold (95% CI, 1.23–6.95) higher in smear-negative tuberculosis than in NTBD subjects. In addition, among the seroreactive subjects, the mean number of reactive proteins was more than twice as high (95% CI, 1.02–4.07]) in smear-positive tuberculosis patients than in NTBD cases. The comparison between smear-negative tuberculosis and NTBD was not statistically significant (data not shown). Thus, immunoproteome reactivity was only sufficiently sensitive to model the differences between tuberculosis and NTBD, whereas reactivity to the tuberculosis-associated proteins was sensitive enough to model differences between all 3 groups (smear-positive tuberculosis, smear-negative tuberculosis, and NTBD). Together, the analyses of the human sera indicate that antibody levels and number of reactive proteins increase with bacillary burden and both parameters of the response vary in concert, as seen in macaques.
DISCUSSION
Given the heterogeneous antibody profiles seen in humans and now also in macaques, tracking antibody responses to individual M. tuberculosis antigens fails to provide a full understanding of the antibody response to M. tuberculosis infection. The correlation between levels of particular antigens and the corresponding specific antibodies likely varies with the antigen considered because antigens may differ in relative immunogenicity and antibodies in relative affinity. Moreover, relative antigen abundance during infection may vary with bacterial growth phase (reviewed in [22]). Although less sensitive than conventional assays such as enzyme-linked immunosorbent assay, our high-throughput platform uniquely allows proteome-scale studies of the antibody response. In the present study, these global measurements revealed novel dynamic characteristics of the antibody response in relation to infection outcome and bacillary burden.
Proteome-scale antibody profiling in infected macaques revealed correlations between antibody response and infection outcome. A transient increase of the antibody response (peaking at around 3 months) was seen both in the latent infection and active disease groups. The early surge is prominent in the latent infection group because antibodies return to quasi-basal levels. In the active disease group, in contrast, a second, stronger, and more stable increase interrupts the descending curve of the early, transient response. That transient, early antibody responses may also be seen in human infection is supported by the observation that contacts of index tuberculosis cases were found to have serum antibodies to particular M. tuberculosis antigens [23], whereas stable latent M. tuberculosis infection in humans is most often seronegative (reviewed in [22]). Furthermore, we find it puzzling that the early peak seen in the active disease and latent groups was absent in the reactivation group, even though the latter group was clinically indistinguishable from the latent group at the time of the transient antibody increase. An intriguing possibility is that stress-induced reactivation occurred in animals that were unable to mount humoral immune responses early after infection.
Integration of macaque and human studies strongly supports correlations between antibody responses and bacillary burden. A vast body of human studies indicates that antibody responses relate to bacterial load because sensitivity of serodiagnostic assays is greater for smear-positive than for smear-negative active tuberculosis (eg, [2, 24, 25]). Our proteome-scale analysis revealed a new aspect of this correlation because reactivity to the immunoproteome was less sensitive to bacillary burden than that to the smaller pool of tuberculosis-associated proteins. Indeed, only reactivity to the tuberculosis-associated proteins differed between high- and low-burden tuberculosis and between either form of tuberculosis and NTBD, indicating that specific protein subsets become preferred targets of the antibody response as disease develops. Moreover, the correlation between antibody responses and bacillary burden in human tuberculosis should help further characterize the course of infection in macaques. That is, the antibody profiles seen in latently infected animals might reflect initial bacillary multiplication, which is subsequently controlled. In contrast, failure of immune control of infection might lead to bacillary multiplication and increased antibody responses in the active disease and reactivation groups. Indeed, that the antibody increase in the reactivation group is clearly delayed relative to the active disease group strongly supports our interpretation of the data. The proposed link among disease activation, bacillary multiplication, and increased antibody responses agrees with reports of increased antibody responses in HIV-positive human subjects progressing to active tuberculosis [4, 26].
The above discussion shows that, even though the antigens targeted by the antibody response may differ between macaques and humans (presumably due to interspecies immunogenetic differences), data obtained in the 2 hosts can be integrated to generate a single, coherent picture of the dynamic properties of the response over time in relation to infection outcome and bacillary burden. Importantly, integration of the macaque and human data leads to at least 3 actionable findings for use in translational research. First, acute infection may be associated with an antibody surge, which is likely transient, suggesting the possibility of diagnosing early infection among contacts of active tuberculosis cases. The lack of this early humoral response—when accompanied by independent proof of infection (eg, by tests assessing cell-mediated responses [27])—may imply greater susceptibility to stress and predict greater reactivation risk. Together, the above considerations support the notion that latent infection comprises a variety of conditions characterized by diverse microbiological, immunopathological, and outcome features [28]. Second, increased antibody responses in latently infected individuals should be a strong indicator of preclinical tuberculosis. Third, proteome-scale work can help identify proteins that have serodiagnostic potential for the entire spectrum of bacillary burden. A caveat is that cases at the high end of this spectrum would be diagnosed more accurately than those at the low end of the spectrum because the regression coefficients in Table 3 are lower and the hurdle models are less sensitive for smear-negative tuberculosis than for smear positive tuberculosis. In conclusion, greater understanding of the antibody response to M. tuberculosis infection afforded by the global measurements reported here supports current efforts of antibody biomarker discovery and reveals new potential applications.
Supplementary Data
Supplementary materials are available at the Journal of Infectious Diseases online (http://jid.oxfordjournals.org/). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. We thank Karl Drlica and Paul Luciw for critical reading of the manuscript. We gratefully acknowledge the past effort of members of the Flynn group in the work with cynomolgus macaques (infection model development, classification of outcome, and serum collection) and of the investigators in the multisite study “Clinical Suspicion of Tuberculosis” (clinical classification, patient recruitment, and establishment of serum bank).
Financial support. This work was supported by the Foundation of Innovative New Diagnostics and the National Institutes of Health (AI069135 and AI063246 to M. L. G.; AI053731 to A. C.; RR022907 to Paul A. Luciw [University of California, Davis]).
Potential conflicts of interest. All authors: No reported conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.World Health Organization. Global tuberculosis control: surveillance, planning, financing. Geneva, Switzerland: World Health Organization; 2010. [Google Scholar]
- 2.Bothamley G, Gennaro ML. The antibody response to infection with Mycobacterium tuberculosis. In: Kaufmann SHE, Britton WJ, editors. Handbook of tuberculosis. Immunology and cell biology. Weinheim, Germany: Wiley-VHC Verlag GmbH & Co. KGaA; 2008. pp. 227–44. [Google Scholar]
- 3.Kunnath-Velayudhan S, Salamon H, Wang HY, et al. Dynamic antibody responses to the Mycobacterium tuberculosis proteome. Proc Natl Acad Sci USA. 2010;107:14703–8. doi: 10.1073/pnas.1009080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gennaro ML, Affouf M, Kanaujia GV, Brusasca PN, Mangura B, Reichman L. Antibody markers of incident tuberculosis among HIV-infected adults in the USA: a historical prospective study. Int J Tuberc Lung Dis. 2007;11:624–31. [PubMed] [Google Scholar]
- 5.Singh KK, Dong Y, Belisle JT, Harder J, Arora VK, Laal S. Antigens of Mycobacterium tuberculosis recognized by antibodies during incipient, subclinical tuberculosis. Clin Diagn Lab Immunol. 2005;12:354–8. doi: 10.1128/CDLI.12.2.354-358.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davies D, Liang X, Hernandez J, et al. Profiling the humoral immune response to infection using proteome microarrays: High throughput vaccine and diagnostic antigen discovery. Proc Natl Acad Sci USA. 2005;102:547–52. doi: 10.1073/pnas.0408782102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Capuano SV, 3rd, Croix DA, Pawar S, et al. Experimental Mycobacterium tuberculosis infection of cynomolgus macaques closely resembles the various manifestations of human M. tuberculosis infection. Infect Immun. 2003;71:5831–44. doi: 10.1128/IAI.71.10.5831-5844.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flynn JL, Capuano SV, Croix D, et al. Non-human primates: a model for tuberculosis research. Tuberculosis. 2003;83:116–8. doi: 10.1016/s1472-9792(02)00059-8. [DOI] [PubMed] [Google Scholar]
- 9.O'Connor SL, Blasky AJ, Pendley CJ, et al. Comprehensive characterization of MHC class II haplotypes in Mauritian cynomolgus macaques. Immunogenetics. 2007;59:449–62. doi: 10.1007/s00251-007-0209-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greene JM, Wiseman RW, Lank SM, et al. Differential MHC class I expression in distinct leukocyte subsets. BMC Immunol. 2011;12:39. doi: 10.1186/1471-2172-12-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kennedy RC, Shearer MH, Hildebrand W. Nonhuman primate models to evaluate vaccine safety and immunogenicity. Vaccine. 1997;15:903–8. doi: 10.1016/s0264-410x(96)00277-0. [DOI] [PubMed] [Google Scholar]
- 12.Wiseman RW, Karl JA, Bimber BN, et al. Major histocompatibility complex genotyping with massively parallel pyrosequencing. Nature Medicine. 2009;15:1322–6. doi: 10.1038/nm.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burwitz BJ, Pendley CJ, Greene JM, et al. Mauritian cynomolgus macaques share two exceptionally common major histocompatibility complex class I alleles that restrict simian immunodeficiency virus–specific CD8+ T cells. J Virol. 2009;83:6011–9. doi: 10.1128/JVI.00199-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin PL, Rodgers M, Smith L, et al. Quantitative comparison of active and latent tuberculosis in the cynomolgus macaque model. Infect Immun. 2009;77:4631–42. doi: 10.1128/IAI.00592-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cleveland WS. Robust locally weighted regression and smoothing scatterplots. J Am Stat Assoc. 1979;74:7. [Google Scholar]
- 16.Liang K-Y, Zeger S. Longitudinal data analysis using generalized linear models. Biometrika. 1986;73:9. [Google Scholar]
- 17.Koenker R, Bassett G. Regression quantiles. Econometrica. 1978;46:33–50. [Google Scholar]
- 18.Mullahy J. Specification and testing of some modified count data models. J Econometrics. 1986;33:341–65. [Google Scholar]
- 19.Cole ST, Brosch R, Parkhill J, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–44. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 20.Bertholet S, Ireton GC, Kahn M, et al. Identification of human T cell antigens for the development of vaccines against Mycobacterium tuberculosis. J Immunol. 2008;181:7948–57. doi: 10.4049/jimmunol.181.11.7948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sitas F, Newton R, Boshoff C. Increasing probability of mother-to-child transmission of HHV-8 with increasing maternal antibody titer for HHV-8. N Engl J Med. 1999;340:1923. doi: 10.1056/NEJM199906173402414. [DOI] [PubMed] [Google Scholar]
- 22.Kunnath-Velayudhan S, Gennaro ML. Immunodiagnosis of tuberculosis: a dynamic view of biomarker discovery. Clin Microbiol Rev. 2011;24:792–805. doi: 10.1128/CMR.00014-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bothamley GH, Beck JS, Potts RC, Grange JM, Kardjito T, Ivanyi J. Specificity of antibodies and tuberculin response after occupational exposure to tuberculosis. J Infect Dis. 1992;166:182–6. doi: 10.1093/infdis/166.1.182. [DOI] [PubMed] [Google Scholar]
- 24.Steingart KR, Dendukuri N, Henry M, et al. Performance of purified antigens for serodiagnosis of pulmonary tuberculosis: a meta-analysis. Clin Vaccine Immunol. 2009;16:260–76. doi: 10.1128/CVI.00355-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Silva VMC, Kanaujia G, Gennaro ML, Menzies D. Factors associated with humoral response to ESAT-6, 38 kDa and 14 kDa antigens in patients with a spectrum of tuberculosis. Int J Tub Lung Dis. 2003;7:478–84. [PubMed] [Google Scholar]
- 26.Laal S, Samanich KM, Sonnenberg MG, et al. Surrogate marker of preclinical tuberculosis in human immunodeficiency virus infection: antibodies to an 88-kDa secreted antigen of Mycobacterium tuberculosis. J Infect Dis. 1997;176:133–43. doi: 10.1086/514015. [DOI] [PubMed] [Google Scholar]
- 27.Pai M, Riley LW, Colford JM., Jr Interferon-gamma assays in the immunodiagnosis of tuberculosis: a systematic review. Lancet Infect Dis. 2004;4:761–76. doi: 10.1016/S1473-3099(04)01206-X. [DOI] [PubMed] [Google Scholar]
- 28.Barry CE, 3rd, Boshoff HI, Dartois V, et al. The spectrum of latent tuberculosis: rethinking the biology and intervention strategies. Nature Reviews Microbiol. 2009;7:845–55. doi: 10.1038/nrmicro2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.