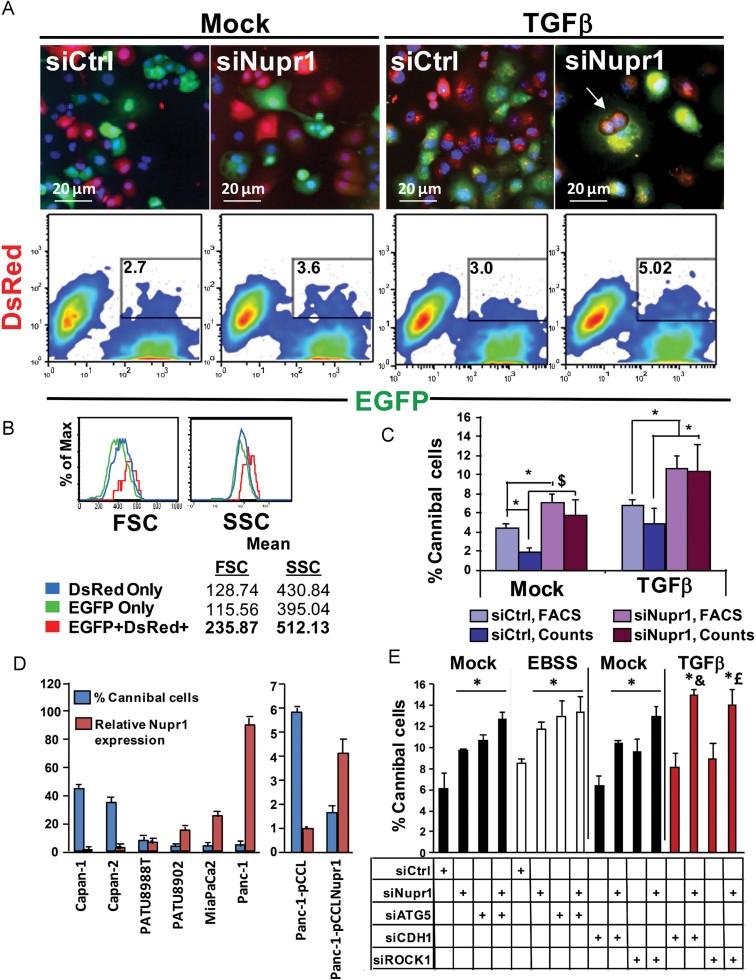
EGFP- and DsRed-expressing Panc-1 cells were mixed in equal numbers and transfected with Nupr1-specific (siNupr1) or control (siCtrl) siRNAs before 48 h of TGFβ treatment. FACS-derived dot plots (bottom) and corresponding fluorescent microscopy pictures (top) illustrate the quantification of cell cannibalism.
Histograms show mean cell size (FSC) and granularity (SSC) of single (GFP only and DsRed only) and cannibal (GFP+DsRed+) TGFβ-treated (48 h) Nupr1-depleted cells as measured by flow cytometry and after exclusion of cell debris.
Histogram shows the microscopic and FACS-based quantification of cannibalism. FACS data are mean of triplicates ±SEM. *p ≤ 0.05, $p ≤ 0.01.
Histogram showing the relative Nupr1 expression (red) in pancreatic cancer cell lines Capan-1, Capan-2, PATU8988T, PATU8902, MiaPaCa2 and Panc-1 compared to Capan-1(left), or Panc-1 transduced with pCCL-Nupr1 or empty pCCL lentiviral vectors (right) as measured by qRT-PCR, and the corresponding % cannibal cells measured by FACS for each cell line (blue).
Like in (A), fluorescent Panc-1 cells were mixed and transfected with the indicated siRNAs, then cultured for 48 h in EBSS medium or in conventional medium with or without TGFβ. Histogram shows the percentage of cannibal cells measured by FACS after 48 h. FACS data are mean of triplicates ± SEM. & and £ are p ≤ 0.05 compared to siCtrl, siCDH1 and siROCK1, respectively.