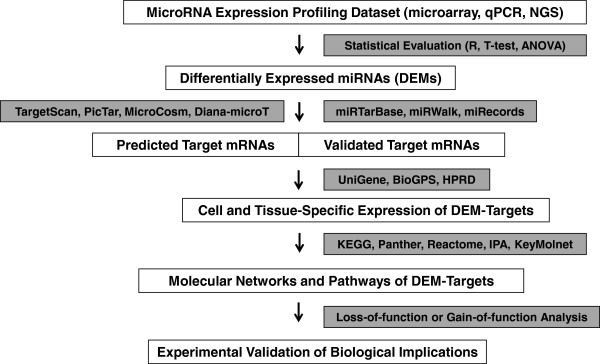
Figure 1.
The workflow of molecular network analysis of microRNA targetome. First, differentially expressed miRNAs (DEMs) among distinct samples and experimental conditions are extracted from microRNA expression profiling datasets based on microarray, qPCR, and next-generation sequencing (NGS) experiments by the standard statistical evaluation. Next, predicted targets and/or validated targets for DEMs are obtained by using target prediction programs, such as TargetScan, PicTar, MicroCosm and Diana-microT 3.0, or by searching them on databases of experimentally validated targets, such as miRTarBase, miRWalk, and miRecords. The expression of DEM targets in the cells and tissues examined is verified by searching them on UniGene, BioGPS, and HPRD. Molecular networks and pathways relevant to DEM targets are identified by using pathway analysis tools, such as KEGG, IPA, and KeyMolnet. Finally, the functionally inverse relationship between miRNAome and targetome is validated by loss-of-function or gain-of-function experiments in an in vitro and/or in vivo model.