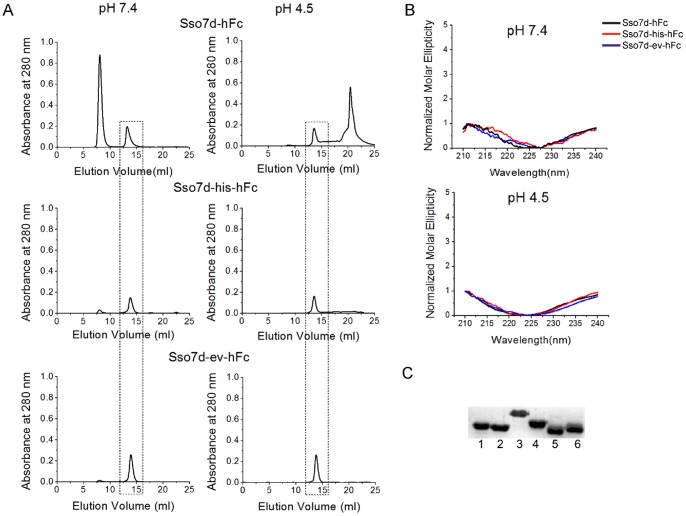
Figure 6. Biophysical characterization of hFc binding Sso7d mutants and western blotting analysis.
(A) Size exclusion chromatography of Sso7d mutants purified by immobilized metal affinity chromatography (IMAC). The dashed box indicates elution peak for Sso7d mutants. Mutants were loaded on the column at a concentration of 2 mg/ml. Molecular weight estimates based on the retention time of Sso7d mutants in the column are consistent with the mutants being present in monomeric form. The other peak corresponds to a minor impurity with higher molar absorptivity than the Sso7d mutants (see Figure S3; SDS-PAGE analysis of fractions corresponding to the other peak do not show any detectable protein). (B) Circular dichroism spectra for Sso7d-hFc, Sso7d-his-hFc and Sso7d-ev-hFc at pH 7.4 and pH 4.5. The spectra at both pH values is essentially the same confirming that there is no change in secondary structure when the pH is lowered from 7.4 to 4.5 (C) Sso7d-hFc recognizes all four hIgG isotypes as well as the deglycosylated form of hIgG, when used as a primary reagent for detection in western blotting analysis. Lane 1: hIgG1, lane 2: hIgG2, lane 3: hIgG3, lane 4: hIgG4, lane 5: hIgG digested with PNGase F, lane 6: undigested hIgG (control). Similar results were observed with Sso7d-his-hFc and Sso7d-ev-hFc (data not shown).