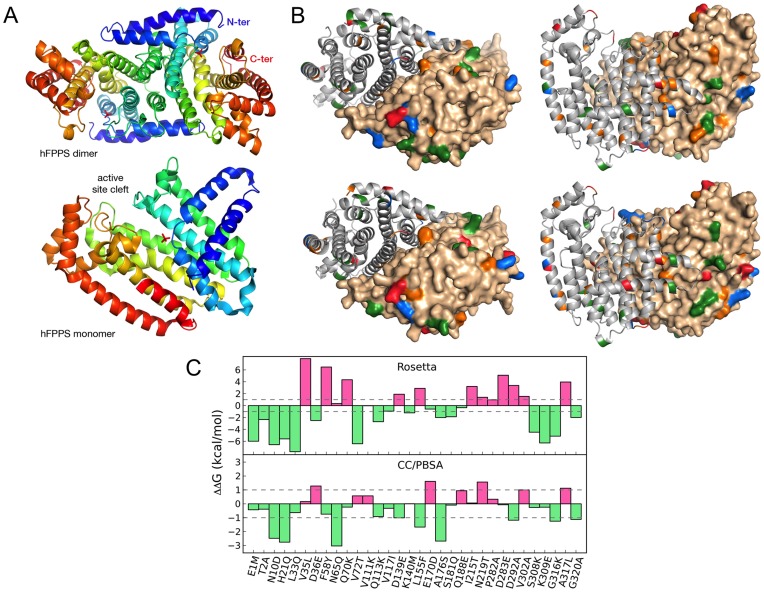
Figure 4. Homology modelling of FPS1S and FPS2 proteins and in silico DDG calculations.
(A) Ribbon representation of the dimeric and monomeric structures of human FPS (PDB 2F7M), which was used to template the threading of Arabidopsis FPS1S and FPS2. The active site cleft is labelled and a bound phosphate ion is shown in sticks. (B) Sequence substitutions between FPS1S (top) and FPS2 (bottom) were mapped onto the ribbon structure (left monomer) or the molecular surface (right monomer) of the homology modeled dimers. Chemical character is colour coded as follows: red, acidic (Asp, Glu); blue, basic (Arg, Lys, His); green, polar (Ser, Thr, Asn, Gln, Tyr); orange, apolar (Met, Phe, Pro, Trp, Val, Leu, Ile, Ala). (C) Histogram of DDG (kcal/mol) upon single-site substitution calculated using Rosetta DDG application (top) or CC/PBSA (bottom). Mutations predicted to occur with a decrease in DDG are coloured green and those expected to increase DDG are coloured pink. Horizontal dashed lines at –1 to +1 kcal/mol bound the neutral area where DDG is supposed to contribute little to the overall stabilization or destabilization of the mutated protein.