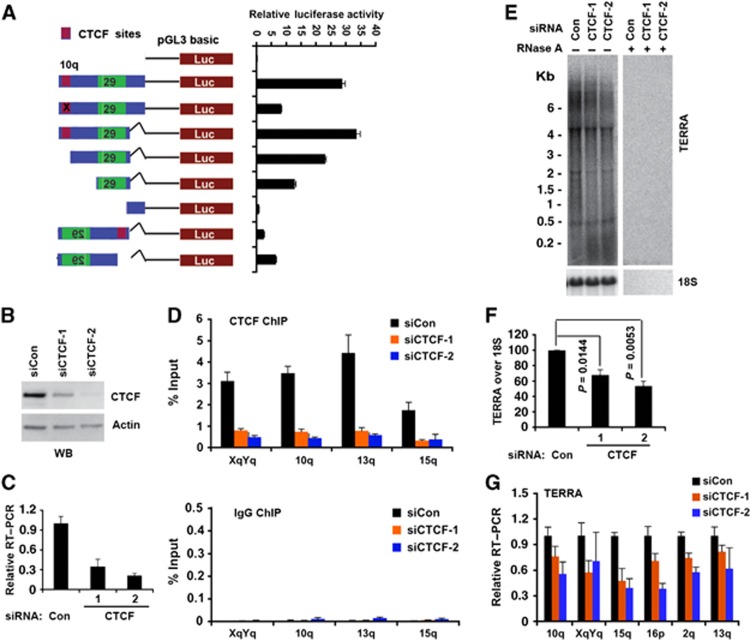
Figure 4.
CTCF function in TERRA transcription. (A) Luciferase reporter constructs containing 10q subtelomere sequence with point mutations in CTCF site (red) or deletion mutations or orientation changes. CTCF-binding site was shown in red and 29 bp element was shown in green. Bar graph represents the average value of relative luciferase activity to Renilla control from three independent transfections (mean±s.d.). (B) Western blot of U2OS cells transfected with siRNA control, siCTCF-1, or siCTCF-2 and assayed with anti-CTCF or anti-actin at 4 days post-transfection. (C) qRT–PCR of U2OS cells transfected with siControl, siCTCF-1, or siCTCF-2 relative to actin mRNA. Relative RT–PCR represents the value calculated by ΔΔCT methods relative to siControl and Gapdh. Bar graph represents the average value from three independent CTCF depletion experiments (mean±s.d.). (D) ChIP-qPCR of CTCF (top panel) or control IgG (lower panel) in U2OS cells transfected with siControl, siCTCF-1, or siCTCF-2 at the CTCF-binding sites in chromosome XYq, 10q, 13q, and 15q presented as percentage of input DNA. (E) Northern blot analysis of TERRA in U2OS cells transfected with siControl, siCTCF-1, or siCTCF-2 with control 18S (lower panel) or RNase A treatment (right panel). Numbers on the left show the position of RNA markers in Kb. (F) Quantification of at least three independent Northern blot assays, a representative is shown in (E). Bar graph represents TERRA signal intensity relative to 18S signal, and relative intensity for siRNA control was set at 100. P-value was calculated by paired two-tailed Student’s t-test (n=3). (G) qRT–PCR of TERRA from individual telomeres at 10q, XYq, 15q, 16q, 2q, and 13q treated with siControl, siCTCF-1, and siCTCF-2 in U2OS cells. Relative RT–PCR represents the value calculated by ΔΔCT methods relative to siControl and Gapdh. Bar graph represents the average value from three independent CTCF depletion experiments (mean±s.d.).