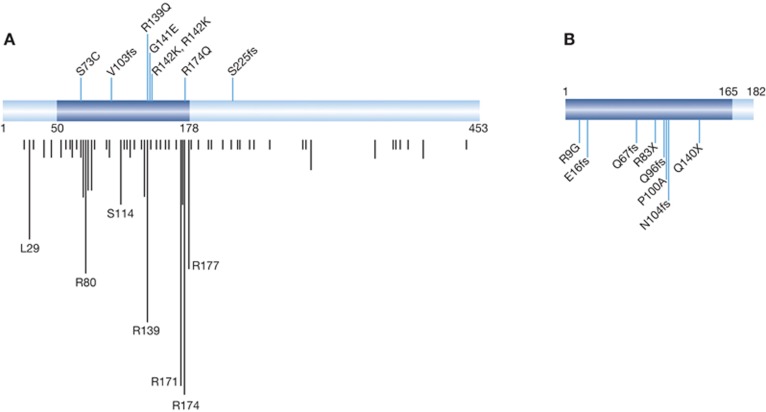
Figure 1.
Mutations in the RUNX1 (A) and CBFB (B) genes. Distribution of eight RUNX1 mutations within RUNX1-coding region as reported in breast cancer. The evolutionally conserved and functionally important Runt domain is highlighted (in blue); all six missense mutations are located within Runt domain, which is responsible for both DNA binding and heterodimerization. Four of the mutations fell within two mutation hot spots (amino-acid position 174 and 139/141/142). Vertical black lines indicate respective mutations reported previously ( http://www.sanger.ac.uk/genetics/CGP/). The length of lines represents cumulative number of mutations. Two frameshift (fs) mutations that result in truncated proteins are as indicated. Notably, all breast cancer-related RUNX1 alterations appear to generate loss-of-function mutants. (B) Distribution of eight CBFB mutations along CBFB/PEBP2β-coding region as reported in breast cancer. RUNX-binding domain is indicated in blue. Six of the mutations are either fs or nonsense, both of which result in non-functional proteins. The remaining two missense mutants (R9G and P100A) are also expected to produce non-functional proteins; R9 and P100 are located within α1 helix and β5 sheet, respectively, both of which form the Runt–CBFβ interface. As a consequence, all of these CBFB genetic changes are likely to result in loss-of-function mutants.