Figure 4.
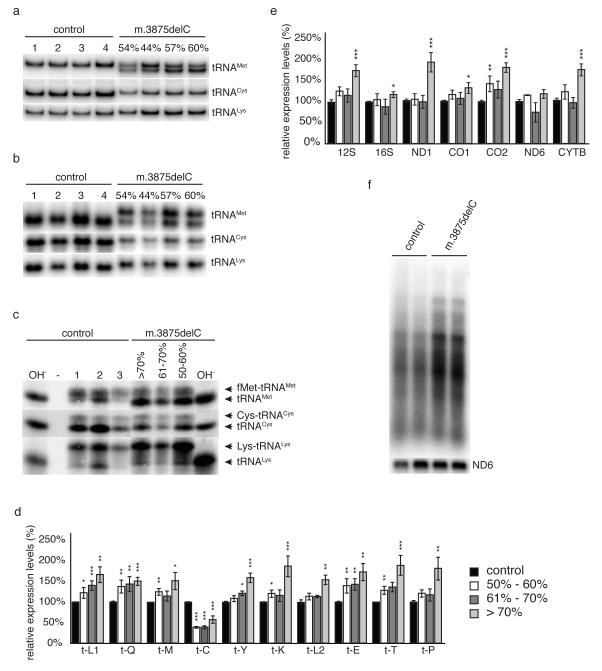
(a-c) Analysis of steady state levels of mitochondrial tRNAs in 16 week old animals with different levels of the m.3875delC mutation of tRNAMet. The percentage of mutated mtDNA refers to the levels in heart. Total RNA was separated by PAGE and blotted and individual tRNAs were detected with radiolabelled probes. (a) Neutral PAGE of total heart RNA demonstrating altered conformation of tRNAMet and decreased levels of tRNACys in mutant animals. (b) Neutral PAGE with non-denatured total heart RNA, demonstrating altered conformation of tRNAMet and tRNACys in mutant animals. (c) Acid-UREA PAGE of total heart RNA extracted under acidic conditions to retain aminoacylation. Base treatment (OH-) of samples prior loading deacylates all tRNAs. (d&e) Relative steady-state levels of mitochondrial (d) tRNAs, (e) mRNAs and rRNAs in hearts from 16 week-old (± 1) mice measured by Northern blot analysis. C57BL/6N (black, n=10-15) m.3875delC mutation at 40-60% (white, n=4-7), 61-70% (dark grey, n=4-7) and >71% (light grey, n=6-11). Levels are normalized to the nuclear encoded 18S rRNA. Error bars show SEM. p<0.05 (*), p<0.01 (**), p<0.001 (***) calculated against C57BL/6N with two-tailed equal variance student t-test. (f) In organello transcription in isolated heart mitochondria from 25 week-old controls and mutant or mice carrying the m.3875delC mutation at >70%.