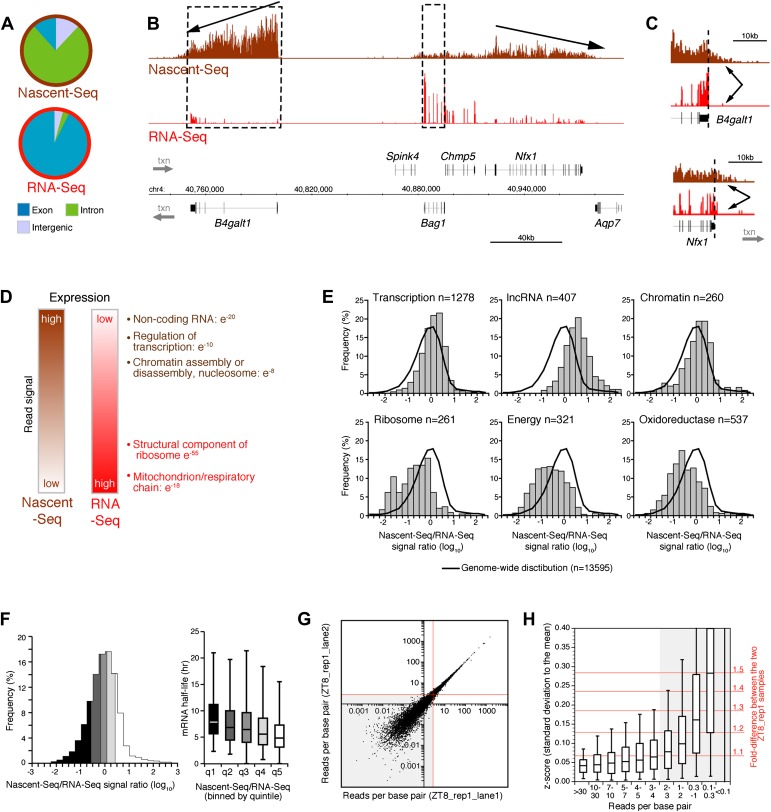
Figure 1. Genome-wide assay of transcription in the mouse liver using Nascent-Seq.
(A): Distribution of high-throughput sequencing signal within introns (green), exons (blue) and intergenic regions (grey) for Nascent-Seq and RNA-Seq datasets. (B): Visualization of Nascent-Seq and RNA-Seq signal at chr4: 40,730,000–41,002,500. Genes above the scale bar are transcribed from left to right and those below the scale bar are transcribed from right to left. Nascent-Seq signal exhibits increased intron signal and a 5ʹ to 3ʹ gradient signal (arrow). Moreover, differences between Nascent-Seq signal and RNA-Seq signal are observed for many genes (e.g., Bag1 and B4galt1). (C): Nascent-Seq signal (brown), but not RNA-Seq signal (red), extends past the annotated 3ʹend of the genes B4galt1 and Nfx1. (D): Gene ontology of genes with high Nascent-Seq and low RNA-Seq signals (and inversely) is indicative of RNA with short or long half-lives, respectively (see ‘Materials and methods’ for details). (E): Distribution of the Nascent-Seq/RNA-Seq signal ratio for the classes of genes enriched in (D). (F): Nascent-Seq/RNA-Seq signal ratio significantly correlates with mRNA half-lives (values from Sharova et al., 2009), and genes with high ratio display shorter half-lives and inversely. (G) and (H): Strategy used to determine the gene signal cut-off threshold used in our analysis. Variation of gene signal coming from the sequencing of a Nascent-Seq library (G; ZT8, replicate 1) sequenced in two Illumina flow-cell lanes was assessed by calculating the z-score (H). Less than 5% of the genes with a read per base pair superior to three exhibit a 1.3-fold gene signal variation. See ‘Materials and methods’ for more details.