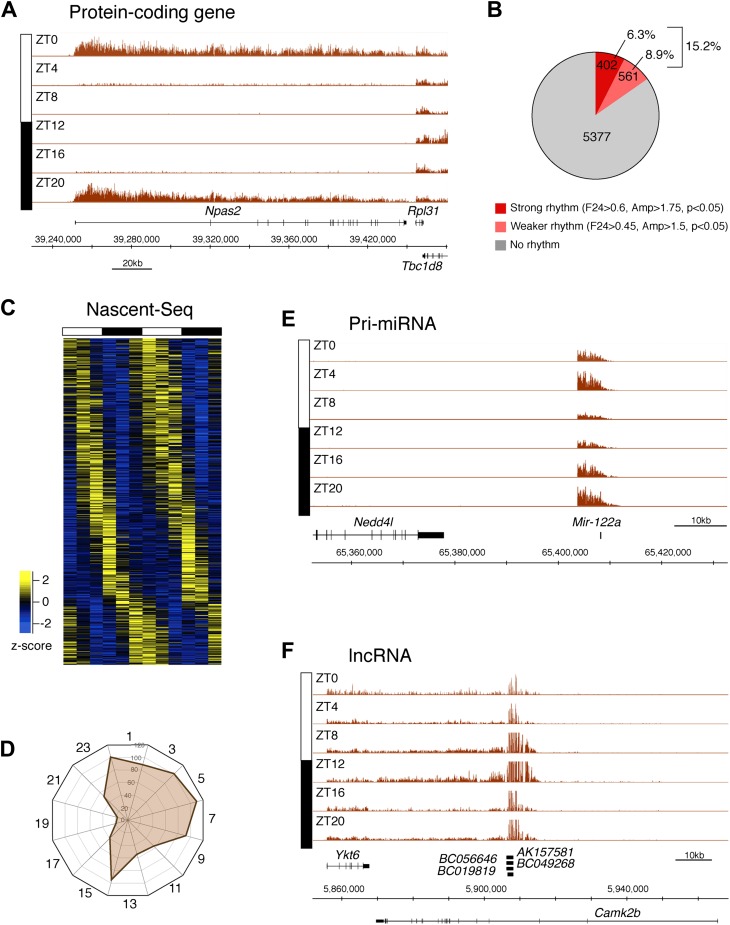
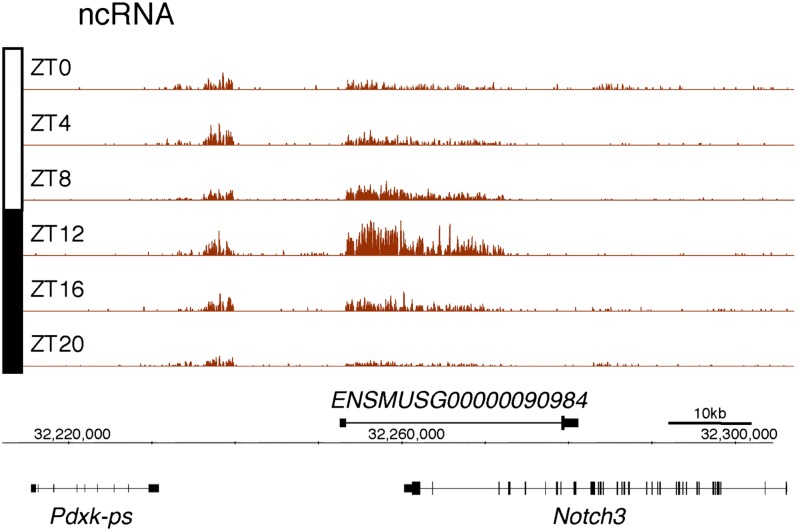
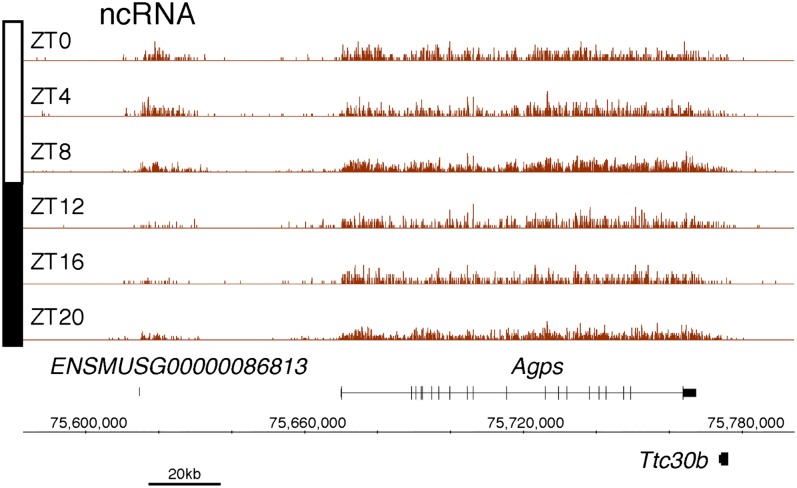
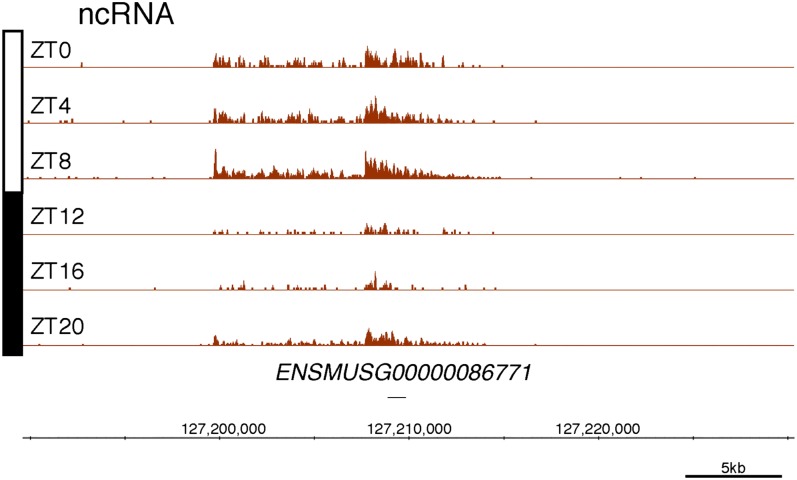
Figure 2. Genome-wide analysis of rhythmic transcription in the mouse liver.
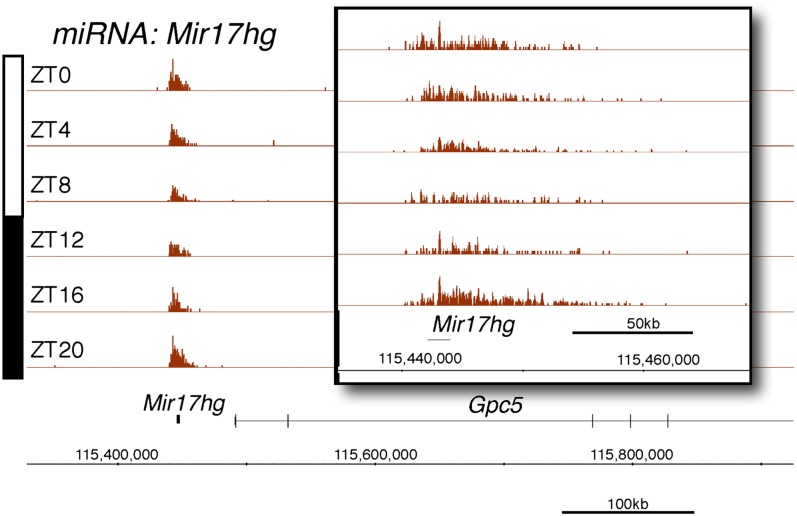
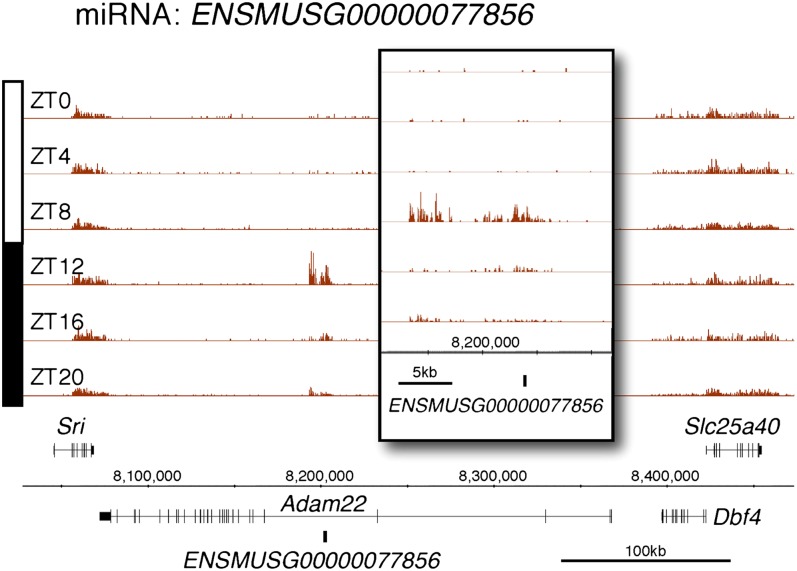
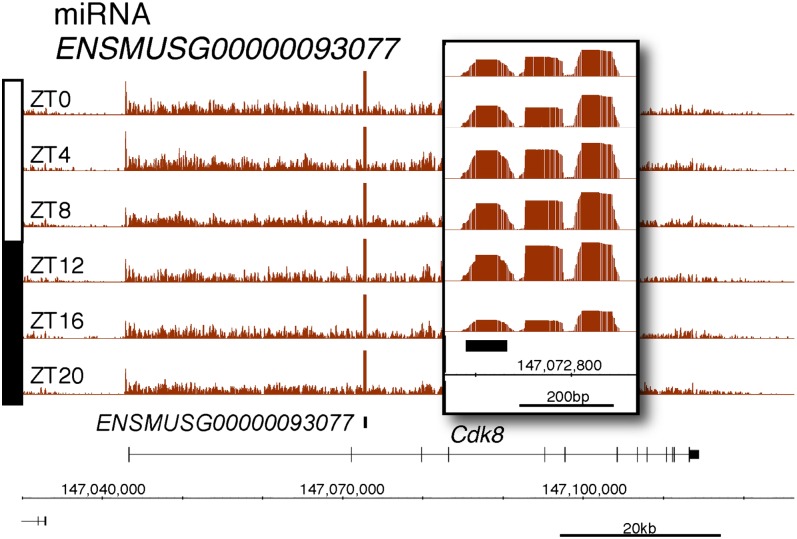
(A): Visualization of Npas2 Nascent-Seq signal at six time points of the light:dark cycle (first replicate). Npas2 Nascent-Seq signal is rhythmic and peaks at ZT20-ZT0, contrary to the signal within the adjacent gene Rpl31. (B): Quantification of the number of genes that are rhythmically transcribed in the mouse liver. Genes with more than three reads per base pair for at least one time point were included for the analysis. Genes are considered to be rhythmically transcribed if signal amplitude (Amp) is greater than 1.5, if signals for the 12 time points follow a sinusoid curve (F24 > 0.45) and if the F24 value is in the top 5% of all F24 values calculated after time points were permutated 10,000 times (p<0.05). A rhythm was considered to be strong (dark red) if F24 > 0.6 and Ampl > 1.75. (C): Heatmap representation of Nascent-Seq signal for the 963 genes that are rhythmically transcribed in the mouse liver. High expression is displayed in yellow (z-score > 1), low expression in blue (z-score < 1). (D): Expression phase of rhythmically expressed nascent RNA (n = 936) was separated by bins of 2 hr. Analysis of their distribution reveals that fewer genes are transcribed at ZT16-20. (E) and (F): Rhythmic Nascent-Seq signal was detected for many precursors of non-coding RNAs such as pri-miRNA (d, pri-miR122a) and long non-coding RNA(e, lin-ncRNAs BC019819, AK157581, BC049268, BC056646).
DOI: http://dx.doi.org/10.7554/eLife.00011.004