Abstract
Label-retaining cells (LRCs) have been proposed to represent adult tissue stem cells. LRCs are hypothesized to result from either slow cycling or asymmetric cell division (ACD). However, the stem cell nature and whether LRC undergo ACD remain controversial. Here, we demonstrate label-retaining cancer cells (LRCCs) in several gastrointestinal (GI) cancers including fresh surgical specimens. Using a novel method for isolation of live LRCC, we demonstrate that a subpopulation of LRCC is actively dividing and exhibits stem cells and pluripotency gene expression profiles. Using real-time confocal microscopic cinematography, we show live LRCC undergoing asymmetric nonrandom chromosomal cosegregation LRC division. Importantly, LRCCs have greater tumor-initiating capacity than non-LRCCs. Based on our data and that cancers develop in tissues that harbor normal-LRC, we propose that LRCC might represent a novel population of GI stem-like cancer cells. LRCC may provide novel mechanistic insights into the biology of cancer and regenerative medicine and present novel targets for cancer treatment.
Keywords: Adult stem cells, Cancer stem cells, Self-renewal, Asymmetric cell division, Cairns immortal strand hypothesis, Liver
Introduction
Label-retaining cells (LRCs) are identified by exposing cells to nucleotide analogs such as bromodeoxyuridine (BrdU) and a chase period without nucleotide analogous. The DNA labels (nucleotide analogous) dilute with each cell division to eventual an undetectable level [1–6]. Data suggest that LRCs are adult tissue stem cells [3–10]. It has been proposed that LRCs are the result of either relative quiescence/slow cycling [3] or asymmetric cell division with nonrandom chromosomal cosegregation (ACD-NRCC) [1, 2, 11]. Recently, several studies have suggested that LRCs are actively dividing, mitigating the slow-cycling hypothesis [4–6, 12–16]. However, the stem cell nature of LRC has been questioned [17, 18].
The concept of ACD-NRCC was introduced by Cairns [11]. It is one possible method by which stem cells divide asymmetrically and self-renew. ACD-NRCC suggests that each chromosome in some stem cells contains one DNA strand that is conserved throughout multiple ACDs (Fig. 1A). By maintaining these DNA template strands within the daughter stem cell, stem cells could avoid accumulation of mutations from replication errors. It is a potential mechanism by which replication errors are preferentially segregated into the daughter cell destined to differentiate and eventually be eliminated like most mature epithelial cells [1, 2, 4–6, 12–16, 19]. However, other investigators could not confirm the existence of ACD-NRCC or LRC [17, 20–23]. The question whether LRCs are generated by ACD-NRCC versus slow cycling remains highly controversial [18].
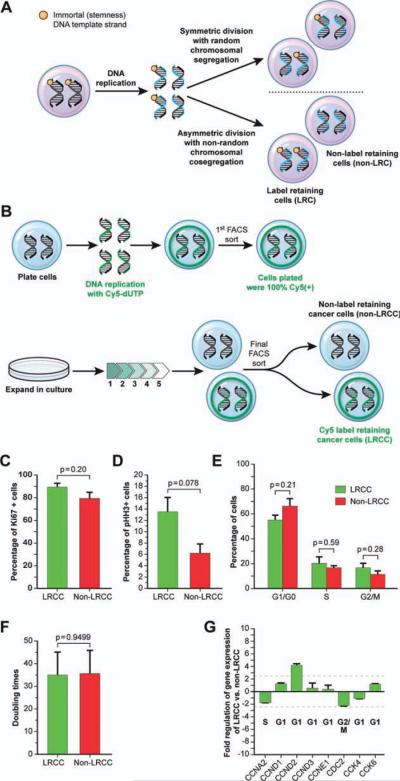
Figure 1.
A subpopulation of LRCC is not quiescent and undergoes active cell division. (A): Asymmetric cell division (ACD) with nonrandom chromosomal cosegregation. Each chromosome in some stem cells contains one template DNA strand that is conserved throughout multiple ACDs (orange mark) and preferentially segregates into the daughter stem cell. (B): Isolation of live LRCC (Materials and Methods and Supporting Information Fig. S1A, S1B). (C–G): Ki67 expression, phospho-histone-3 expression, cell cycle phases, doubling times, and key cell cycle check point genes expression in LRCC versus non-LRCC are not statistically different indicating that LRCCs are not quiescent. Abbreviations: dUTP, 2′-deoxyuridine 5′-triphosphate; FACS, fluorescence-activated cell sorting; LRC, label-retaining cell; LRCC, label-retaining cancer cell; CCNA2, cyclin A2; CCND1, cyclin D1; CCND2, cyclin D2; CCND3, cyclin D3; CCNE1, cyclin E1; CDC2, cell division control protein 2; CDK2; cyclin-dependent kinase 2; CDK4, cyclin-dependent kinase 4; CDK6, cyclin-dependent kinase 6.
To test the cancer stem cells hypothesis [24–32], we developed a novel methodology that enables us to isolate live label-retaining cancer cells (LRCCs). Based on the fact that solid organ cancers are derived from tissues that contain LRC, and that LRCs are thought to be adult tissue stem cells, we tested human cancer cell lines and fresh surgical cancer specimens for the existence of LRCC. We tested whether LRCC possess stem-like properties, and the mechanism by which LRCCs are generated. Finally, based on the cancer stem cell hypothesis, we tested their tumor-initiating capacity in immunocompromised mice.
Here, we show the existence of LRCC in gastrointestinal (GI) cancers. LRCC express proliferation markers, cell cycle checkpoint genes, and a mitotic marker suggesting that LRCCs are not quiescent but rather undergo active cell division. For the first time, to our knowledge, we demonstrate live LRCC undergoing label-retaining ACD with NRCC. Finally, we demonstrate that LRCCs have greater tumor-initiating capacity than non-LRC generating tumors with only 10 cells, and a stem cells gene expression profile. Taken together, these findings suggest that LRCC represent a novel class of common GI cancer stem cells, and as such may provide new insights into the biology of cancer and the stem cell origins of cancer.
Materials and Methods
Fresh Primary Human Cancer Cells and Cancer Cell Lines
Fresh tissue was obtained on National Cancer Institute protocol 09-C-0079. Tumors were harvested and processed into spheroids, transplanted into nude mice once, harvested, and used in this study (Supporting Information Materials and Methods and Table S1).
Ki67 and Phospho-Histone-3 Detection by Fluorescence-Activated Cell Sorting Analysis
Staining for Ki67 (Ki67-FITZ, Dako, Glostrup, Denmark, http://www.dako.com) and phospho-histone-3 (pHH3) (pHH3-Alexa-488, S-10, Cell Signaling, Boston, MA, http://www.cellsignal.com) was done as per manufacturer instructions. Data were acquired on BD FACS aria II and analyzed with FlowJo (Ashland, OR, http://www.flowjo.com, Supporting Information).
Isolation of Live LRCC
Isolation of live LRCC was done as described (Supporting Information Fig. S1A, S1B and Materials and Methods).
Real-Time Confocal Cinematography for the Detection of LRCC Undergoing ACD-NRCC
Cancer cells with Cy-5-labeled DNA were isolated and plated onto collagen-IV-coated slide chambers (Supporting Information Fig. S2 and Materials and Methods). Confocal cinematography imaging was performed on a Zeiss LSM 710 NLO confocal equipped with an environmental chamber (Carl Zeiss, Deutsch-land, Germany, http://www.zeiss.de).
Gene Expression Analysis
Total RNAs were isolated using miRNeasy Mini kit and RNase-Free DNase Set (QIAGEN, Valencia, CA, http://www.qiagen.com). RNA quantification (Nanodrop), quantitative real-time reverse transcription-polymerase chain reaction (qRT-PCR), and Ct value analysis were done for human stem cell, pluripotency, and the wingless-type MMTV integration site family (WNT) pathway SuperArrays in triplicates using 384-well plates with ABI 7900 HT system (Applied Biosystems, Carlsbad, CA, http://www.appliedbiosystems.com) according to the manufacturer's protocol (SABiosciences, Valencia, CA, http://www.sabiosciences.com).
Mouse Xenogeneic Transplantation
LRCC and non-LRCC were isolated after eight cell cycles from the human hepatocellular carcinoma (HCC) cell line (PLC/PRF/5) and fresh primary colorectal cancer cells (CSCL-04-Ke). We transplanted subcutaneously 10 cells with 25% of Matrigel into nude severe combined immunodeficiency (Nude/SCID) mice (n = 40; SHO, Jackson Lab). Mice were tagged with transponders (Bio Medic Data Systems, Inc, Seaford, DE, http://www.bmds.com).
Statistics
All data are presented as the means ± SEM. Statistical differences were evaluated as follows: (a) the statistical significance of observing ACD-NRCC was calculated with the two-tailed p value by the exact binomial test. (b) Fisher's exact test was used to test for significance of tumor-initiating capacity (Supporting Information Materials and Methods).
Results
A Subpopulation of LRCCs Is Not Quiescent and Undergoes Active Cell Division
We developed a novel method that allowed for the isolation of live LRC (Materials and Methods). To test whether LRCC undergo active cell division, we isolated live LRCC and non-LRCC (Fig. 1B) from three HCC cell lines and three surgical specimens (three colon cancers, Supporting Information Materials and Methods). The relative percentages of LRCC ranged from 1.3% to 2.0% (n = 6).
Ki67 is a nonspecific cell cycle marker (G1, S, and G2/M phases). pHH3 is a mitotic marker (Materials and Methods). Fluorescence-activated cell sorting (FACS) analysis revealed that 89.4% ± 3.3% versus 79.2% ± 5.2% of the LRCC and non-LRCC are Ki67 positive (p = .20), respectively (Fig. 1C). Additionally, 13.5% ± 2.5% versus 6.5% ± 1.6% of the LRCC versus non-LRCC are positive for pHH3 (p = .078), respectively (Fig. 1D). These results suggest that there is no difference between the proportion of LRCC and non-LRCC cells undergoing active mitosis. Furthermore, we found that LRCC undergo active cell division: 55.3% ± 3.9%, 20.3% ± 5.4%, and 16.9% ± 3.4% of the LRCC are in G1/G0, S, and G2/M phases, respectively. In comparison to the non-LRCC, there is no difference in the proportion of LRCC that are in G1/G0, S, and G2/M phases, p = .21, p = .59, and p = .28, respectively (Fig. 1E) These results suggest that a subpopulation of LRCC is not quiescent and undergo active cell division.
To validate these findings, we tested the cell cycle duration of LRCC and the non-LRCC. The cell cycle duration of LRCC was 34.9 ± 8.8 hours, and the cell cycle duration of the non-LRCC was 36 ± 9.2 (n = 18, p = .95, Fig. 1F). Finally, we tested and compared LRCC versus non-LRCC for the expression of key cell cycle checkpoint genes. Using qRT-PCR cell cycle array, we show that there is no statistical difference in the expression of all tested genes (cyclin A2, CCNA2; D1, CCND1; D2, CCND2; D3, CCND3; E1, CCNE1; cell division control protein 2, CDC2; cyclin-dependent kinase 2, CDK2; 4, CDK4; and 6, CDK6) between LRCC and non-LRCC (Fig. 1G, n = 18). Interestingly, CCND2, a gene expressed during the mid-G1 and exit from G0–G1 phase, was expressed 4.2 ± 0.2-fold higher in the LRCC than in the non-LRCC. Exit from G0 into the G1 phase is thought to herald stem cells activation.
In summary, using five layers of evidence, we show that a subpopulation of LRCC is actively dividing, mitigating the quiescence/slow-cycling hypothesis in LRCC. These findings suggest that LRCC could undergo ACD-NRCC.
LRCC Undergo ACD with NRCC
To test the alternative hypothesis asking the question whether LRCC undergo ACD-NRCC, we developed a novel method to detect live LRCC (Fig. 2A, Materials and Methods). Cells were grown for one cell cycle in serum-free media and underwent a double-thymidine arrest to increase the probability of cells being synchronously in G1-S phase at the inception of the experiment. Subsequently, we added complete media and allowed DNA synthesis to occur in an environment rich with the DNA nucleotide analog Cy5-dUTP (2′-deoxyuridine 5′-tri-phosphate), as described in Materials and Methods. After incorporation of Cy5-dUTP into the DNA, cells were grown for one more cell cycle in culture. Using FACS, we sorted only Cy5-dUTP-high positive cancer cells with >99% purity. Cy5-dUTP-positive cancer cells were then placed on collagen-IV-coated chamber slides, and their nuclei were labeled with the vital stain Cyto9. Subsequently, we initiated continuous confocal microscopic cinematography of live cells undergoing cell divisions. In Figure 2B, 2C, we show one such representative ACD-NRCC. The still pictures in Figure 2B, 2C, were taken from a continuous video where at time t = 0 minute, one can see a single cell with a single nucleus containing DNA labeled with Cy5-dUTP (Fig. 2B, green). Following the same cell, at time t = 210 minutes, one can observe one cell with two nuclei during mitosis; however, here, only one of the nuclei contains Cy5-dUTP-labeled DNA (Fig. 2C and Supporting Information Video S1). At time t = 600 minutes, one can observe two cells: one with Cy5-dUTP-labeled DNA (Fig. 2B, green and Supporting Information Video S1) and the other with unlabeled DNA (Fig. 2B, blue and Supplemental Video S1). To ascertain that these are not two cells over each other, we used confocal microscopic cinematography to deconstruct the layers (Z stacking) confirming one cell dividing into two. To fully appreciate this phenomenon, we attached a video of live LRCC undergoing ACD-NRCC in real time (Supporting Information Video S1). As far as we know, this is the first time, to our knowledge, that ACD-NRCC is recorded in live cells and in real time. In the first set of experiments, we observed 104 cell divisions in three different experiments, 2/104 of these cells underwent ACD-NRCC. In subsequent experiments (n = 16), the relative proportion of cells undergoing ACD-NRCC was 1.9%–2.7%. LRCC undergoing ACD-NRCC is a rare but statistically significant phenomenon (p = .001, statistics in Materials and Methods).
Figure 2.
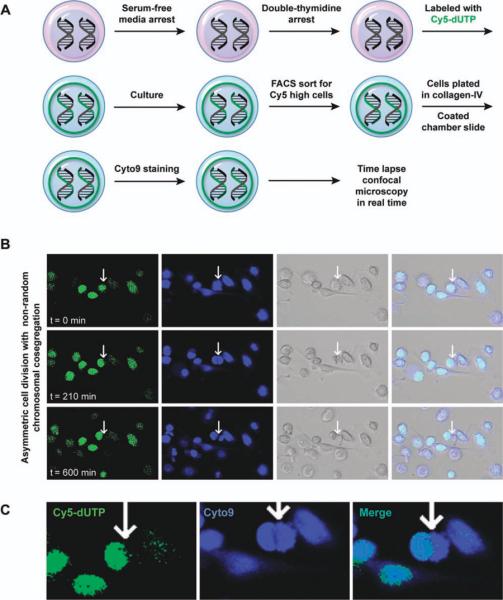
Live label-retaining cancer cell (LRCC) undergoes asymmetric cell division with nonrandom chromosomal cosegregation (ACD-NRCC) in real time. (A): Materials and methods to detect LRCC undergoing ACD-NRCC (Materials and Methods). (B): Confocal microscopic cinematography. Following a single cell: at t = 0 minute, single cell labeled with Cy5 (green). t = 210 minutes, single cell, two nuclei (Blue-cyto9), and only one is retaining the Cy5. t = 600 minutes, two cells, two nuclei, and only one retains Cy5. Images were confirmed using confocal Z-stack images. (C): LRCC undergoing ACD-NRCC, showing a cell (t = 210) in a middle of mitosis, two nuclei within the same cytoplasmic space; however, only one nucleus retains the Cy5 DNA label. (Supporting Information Fig. S2 and Video S1). Cyto9 is a vital dye; Cy5 is a fluorescent dye. Abbreviations: dUTP, 2′-deoxyuridine 5′-triphosphate; FACS, fluorescence-activated cell sorting.
LRCC Exhibit Greater Tumor-Initiating Capacity than Non-LRCC
To further understand the biological implications and the potential stem cell nature of LRCC, we tested the tumor-initiating capacity of LRCC (Materials and Methods). We isolated live LRCC and non-LRCC from one HCC cell line (PLC/PRF/5) and fresh cancer cells from a surgical specimen (CSCL-04-Ke derived from colorectal cancer). All in vivo experiments were done in a blinded fashion. Moreover, mice were scrambled blindly within and among cages, and we used coded electronic transponders to track the mice. All experiments were terminated at 16 weeks. The sealed envelope containing the blinding code was opened in the presence of all involved. We transplanted 10 cells into 20 Nude/SCID mice per each group (LRCC group and non-LRCC group). We found that LRCC exhibited superior tumor-initiating capacity when compared with non-LRCC: 14/20 versus 2/20 of the mice generated tumors (p = .0005, Fisher's exact test). The LRCC generated faster and larger tumors than the non-LRCC, 8 weeks versus 14 weeks (Fig. 3).
Figure 3.
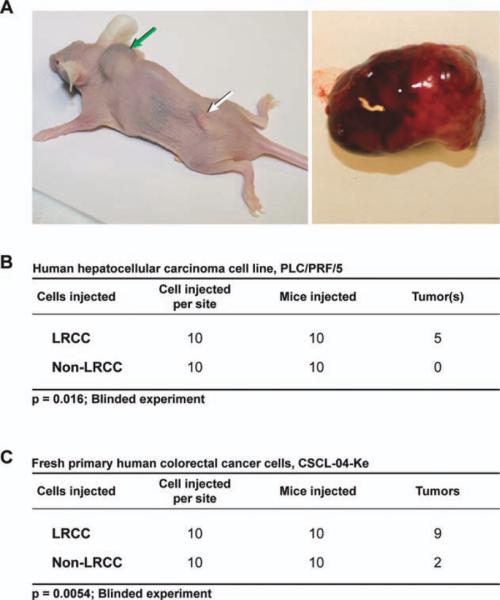
LRCCs exhibit greater tumor-initiating capacity than non-LRCC. (A): Tumors generated by only 10 LRCC (green arrow). Transponders used in the blinding procedure (white arrow). A tumor is demonstrated on the right. (B, C): Two cell lines were used: PLC/PRF/5 (hepatocellular carcinoma) and fresh colorectal cancer (CSCL-04-Ke). In a blinded experiment, 14/20 versus 2/20 of the animals grew tumors from only 10 cells (p = .0005, Fisher's exact). Abbreviation: LRCC, label-retaining cancer cell.
Stem Cells and Pluripotency Gene Expression Profiling of LRCC
To gain further understanding of the potential stem cell nature of LRCC, we isolated live LRCC and non-LRCC and compared their gene expression profiles (Materials and Methods). We performed qRT-PCR SuperArray analysis: WNT (84 genes), stem cells (84 genes), and pluripotency (11 genes). We analyzed three HCC cell lines and three freshly isolated colon cancers from surgical specimens (n = 18).
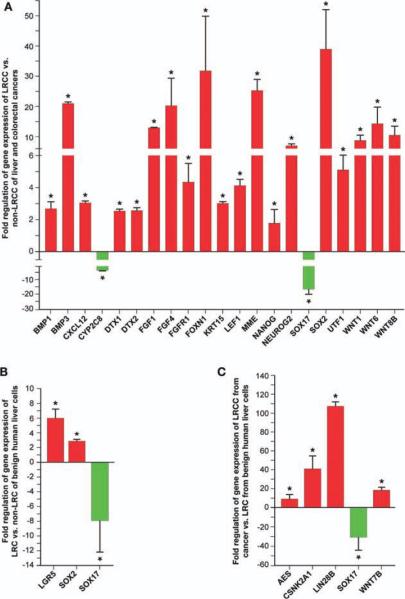
Of the 179 genes analyzed, 21 genes were differentially expressed (Fig. 4A). Two genes were downregulated: SRY (sex determining region Y)-box 17 or SOX17 (−16.3 6 ± 3.3-fold, p = .013) and cytochrome P450 family 2 subfamily C polypeptide 8 or CYP2C8 (−3.9 ± 0.3-fold, p = .021). SOX17 knockdown induced dedifferentiated state [33]. Denovo expression of SOX17 induced hyperplasia and differentiation [34]. CYP2C8 can induce differentiation in some stem cells. Thus, down regulation of SOX17 and CYP2C8 is consistent with a stem-like cell profile.
Figure 4.
Stem cells and pluripotency gene expression profiling of LRCC. We performed quantitative real-time (qRT)-PCR SuperArray analysis of human stem cells (84 genes), pluripotency (11 genes), and Wnt (84 genes) pathway genes. (A): Gene expression profiling of LRCC versus non-LRCC. (B): Gene expression profiling of LRC versus non-LRC from benign liver cells. (C): Gene expression profiling of LRCC versus LRC. A comparison between cancer and benign LRCs (genes in red are upregulated, green downregulated). * indicates p < .05. Abbreviations: AES, amino-terminal enhancer of split; BMP, bone morphogenetic protein; CSNK, casein kinase; CXCL, chemokine (C-X-C motif) ligand; CYP, cytochrome P450; DTX, deltex homolog; FGF, fibroblast growth factor; FOXN, forkhead box N; KRT, keratin; LEF, lymphoid enhancer-binding factor; LGR, leucine-rich repeat containing G protein-coupled receptor; LIN, lin-28 homolog; LRC, label-retaining cell; LRCC, label-retaining cancer cell; MME, membrane metallo-endopeptidase; NANOG, nanog homeobox; NEUROG2, neurogenin 2; SOX, SRY (sex determining region Y)-box; UTF, undifferentiated embryonic cell transcription factor; WNT, Wingless-type MMTV integration site family.
Nineteen genes were upregulated by LRCC (Fig. 4A). SOX2 (38.9 ± 13.1-fold, p = .035) is a transcription factor essential for self-renewal, maintenance of undifferentiated state, and pluripotency [35, 36]. Nanog homeobox (NANOG) and undifferentiated embryonic cell transcription factor 1 (UTF1), both associated with pluripotency and self-renewal (2.2 ± 0.1-fold, p = .0040 and 5.1 ± 0.9-fold, p = .072). We show that LRCC express six reported pluripotency genes (octamer-binding protein 3/4, OCT3/4; SOX2; NANOG; v-myc myelocytomatosis viral oncogene homolog, c-MYC; lin-28 homolog A, LIN28; and Kruppel-like factor 4, KLF4) [35, 36] but significantly upregulated only three: NANOG, SOX2 and LIN28. Expression of these six genes with highly unusual upregulation of SOX2 (38.9 ± 13.1-fold, p = .035, Fig. 4A) and LIN28 (107.5 ± 4.4-fold, p = .0089, Fig. 4C) further supports the stem cell nature of LRCC.
WNT signaling plays an important role in pluripotency [37] and stem cells function [38]. We found that WNT6, WNT8A and WNT1 were upregulated in LRCC by 14.4 ± 5.7 (p = .0033), 10.5 ± 3.0 (p = .033), and 8.6 ± 2.1 (p = .013) folds, respectively (Fig. 4A). LEF1 (4.1 ± 0.4-fold, p = .047) is a WNT signaling transcription factor.
Several WNT pathway target genes and genes associated with self-renewal and stem cell maintenance were upregulated by the LRCC. Forkhead box N1 (FOXN1) was upregulated by 32.1 ± 17.7 (p = .0040) folds (Fig. 4A). FOXN1 is a stem cell transcription factor associated with embryonic development [38–40]. The mRNAs encoding for the notch 1 (NOTCH1) and notch 2 (NOTCH2) agonists, deltex homolog 1 and 2 (DTX1 and DTX2) [41, 42], are increased by 2.5 ± 0.1 (p = .025) and 2.6 ± 0.2 (p = .00043) folds, respectively (Fig. 4A). Neurogenin 2 (NEUROG2) is associated with self-renewal and stem cell maintenance, 7.0 ± 0.6-fold (p = .020). Membrane metallo-endopeptidase or MME (Neprilysin, CD10) was found to be upregulated by the LRCC by 25.3 ± 3.6 (p = .012) folds. It is an important regulator of cell migration and metastasis [43, 44]. Keratin 15 or KRT15 (3.0 ± 0.1-fold, p = .037) is a type I cytokeratin and is highly expressed by epithelial progenitor cells [45].
Fibroblast growth factor 4 (FGF4) (20.3 ± 9.2-fold, p = .0011) is having important role in embryonic development. Expression of FGF4 induces epithelial hyperplasia and inhibition of apoptosis [46]. FGF4 is restricted in vitro to embryonal carcinoma, and it is repressed during differentiation [47]. FGF4 is required for human embryonic stem cell pluripotency and is regulated by SOX2 and OCT3/4 [37, 48, 49]. FGF1 (13.0 ± 0.1, p = .048) is a modifier of cell migration and organogenesis. It interacts with chemokine (C-X-C motif) receptor 4 (CXCR4) to regulate stem cell migration. We found that bone morphogenetic protein 1 and 3 (BMP1 and BMP3) expression is increased by 2.7 ± 0.4 (p = .0021) and 21.0 ± 0.6 (p = .0029) folds, respectively (Fig. 4A). BMP1 is an important negative regulator of BMP2, BMP4, and BMP7 involved in pluripotency and self-renewal [37, 50]. Lastly, chemokine (C-X-C motif) ligand 12 (CXCL12) or stromal cell-derived factor 1 (3.1 ± 0.1-fold, p = .00048), SDF1, is an important chemokine during embryogenesis regulating stem cells migration from the liver to the bone marrow [51].
We compared the stem cells gene expression profile of benign (noncancer) LRCs versus non-LRCs from two different noncancer liver cell lines. We show that LRC differ from non-LRC by upregulating leucine-rich repeat containing G protein-coupled receptor 5 or LGR5 (6.2 ± 1.1, p = .00022) and SOX2 (2.9 ± 0.2, p = .00033) and downregulating SOX17 (−7.9 ± 4.3, p = .029) (Fig. 4B). LGR5 is a marker of GI stem cells [52]. Finally, we studied the differences between liver LRCC and normal adult liver LRC. Of 179 genes studied, only five were differentially expressed (Fig. 4C): The WNT pathway associated genes amino-terminal enhancer of split or AES (10.1 ± 5.1, p = .044), casein kinase 2 alpha 1 polypeptide or CSNK2A1 (43.6 ± 14.6, p = .00044), WNT7B (19.6 ± 3.4, p = .014), and SOX17 (−33.3 ± 14.8, p = .0037). Interestingly, LIN28B (107.5 ± 4.4, p = .0089) was highly overexpressed by the LRCC [36, 53, 54].
Taken together, our data suggest that LRCCs have gene expression profile consistent with stem-like phenotype. To integrate these results, we used the Ingenuity pathway analysis software to generate a stem cell pathway map for LRCC (Fig. 5).
Figure 5.
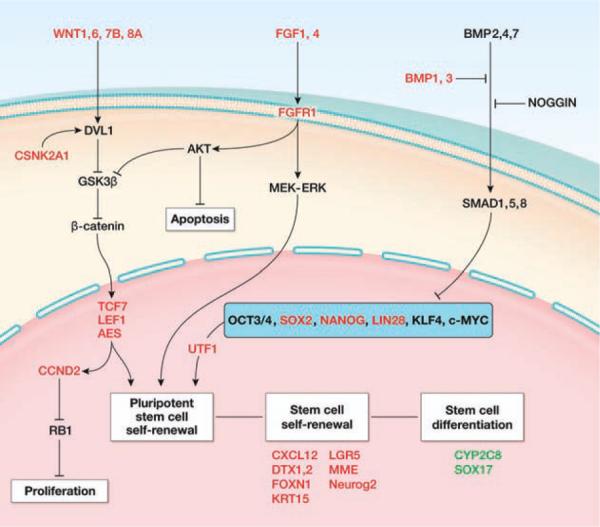
Stem cell pathways analysis for label-retaining cancer cell (LRCC). Using our gene expression data and the ingenuity pathway analysis platform, we show that LRCC upregulates known stem cells, pluripotency, and Wnt pathway genes and downregulates genes associated with gastrointestinal differentiation and apoptosis suggesting a stem cell gene expression profile. Abbreviations: AES, amino-terminal enhancer of split; AKT, v-akt murine thymoma viral oncogene homolog; BMP, bone morphogenetic protein; CCND, cyclin D; CSNK, casein kinase; CXCL, chemokine (C-X-C motif) ligand; CYP, cytochrome P450; DTX, deltex homolog; DVL, dishevelled; FGF, fibroblast growth factor; FOXN, fork-head box N; GSK, glycogen synthase kinase; KLF, Kruppel-like factor; KRT, keratin; LEF, lymphoid enhancer-binding factor; LGR, leucine-rich repeat containing G protein-coupled receptor; LIN28, lin-28 homolog; MEK-ERK, mitogen-activated protein kinase kinase-MAP kinase; MME, membrane metallo-endopeptidase; MYC, v-myc myelocytomatosis viral oncogene homolog; NANOG, nanog homeobox; NEUROG, neurogenin; NOGGIN, noggin; OCT, octamer-binding protein; RB, retinoblastoma; SMAD, mothers against DPP homolog; SOX, SRY (sex determining region Y)-box; TCF, T-cell specific transcription factor; UTF, undifferentiated embryonic cell transcription factor; WNT, Wingless-type MMTV integration site family.
Discussion
Although, several investigators using membrane dyes demonstrated the existence of quiescent non-DNA LRCs in rodents, little evidence exists to suggest the existence of nonquiescent DNA LRCs in human cancers (LRCC) [12]. Demonstrating nonquiescent DNA LRCCs in GI cancers is shown here for the first time, to our knowledge. It gives further credence to the hypothesis that cancers, similarly to adult tissue, might be driven by stem-like cells.
The existence of ACD-NRCC has been questioned. Among several reasons for the controversy was the inability to reproducibly test live LRC or LRCC. Here, we show live LRCC undergoing ACD-NRCC in real time. Potentially, the existence of LRCC undergoing ACD-NRCC may pave the way for novel strategies to target cancer via targeting the mechanisms underlying LRCC. Furthermore, using similar studies, it may provide novel understanding into adult tissue stem cells (LRC) and regenerative medicine.
Isolation of cancer stem cells has been based mostly on cell-surface markers or the side population. More recently Pine et al. demonstrated the existence of cells that undergo ACD in lung cancer [12]. However, their observation was done on fixed cells, and thus they were unable to test whether these cells function like stem cells. Our method uses the ability for stem cells to retain DNA labels. Since LRCCs were identified in diverse GI cancers and most of GI cancers develop in tissues known to harbor LRC, it is conceivable that the property of DNA label retaining could be used to study a potential common stem-like cancer cells in GI cancers. This is an exciting prospect as it may provide a common platform for the comparison of malignant transformation among diverse stem-like cells. This can provide invaluable insights for the development of novel cancer therapeutics against LRCC. Additionally, it may provide a common platform to study the differences between normal adult tissue stem cells (LRC) and stem-like cancer cells (LRCC), providing further insights into carcinogenesis and potentially adult tissue regeneration.
In view of the fact that LRCC undergo active cell division, the question of relative resistance to chemotherapy must be explained differently. Clearly, quiescence can explain resistance of cancer stem cells to chemotherapy. However, if LRCCs undergo active cell division there must be other mechanisms to protect them from chemotherapy. Further investigation of such mechanisms could be the base for a novel approach for anticancer drug developments.
Here we show, for the first time, to our knowledge, that putative HCC and colorectal cancer stem cells, that is, LRCC can generate tumors with only 10 cells. The LRCCs have superior tumor-initiating capacity than the non-LRCC (p = .0005). Previous studies using different cancer stem cells' markers (side population, CD133, CD44/CD24/EpCam) demonstrated tumor-initiating capacity consistently always with more than 10 cells. However, there is fundamental variability among studies in terms of conditions. The LRCC from established cell lines and fresh tumors generated large tumors within 8 weeks using only 10 cells (FACS technique). Thus, based on their ability to generate tumors, LRCC should be considered as putative novel stem-like cancer cells or tumor-initiating cells.
A potential drawback of our methodology is the introduction of modified nucleotides into the cells. However, since the non-LRCCs are derived from cells that underwent introduction of modified nucleotides, like the LRCC, but did not retain DNA label, it is unlikely that the introduction of modified nucleotides was the source of LRCC behaving like stem cells.
In conclusion, using multiple lines of evidence, we demonstrated that LRCC possess stem cells' traits. We showed that LRCC undergo ACD-NRCC, a property suggested previously only to stem cells. We demonstrated that LRCCs have exquisite ability to initiate tumors with only 10 cells, a property associated with stem-like cancer cells. We demonstrate that LRCCs when compared with non-LRCCs have stem cells' gene expression profile. In particular, LRCCs express all six human genes used to generate induced pluripotent stem (iPS) cells (OCT3/4, SOX2, NANOG, c-MYC, LIN28 and KLF4). Expression of these six genes with highly unusual upregulation of SOX2 (38.9 ± 13.1-fold, p = .035) and LIN28 (107.5 ± 4.4-fold, p = .0089) is highly suggestive of stem cell gene expression profile. We propose that the LRCC could be common subpopulation of novel stem-like cancer cells or tumor-initiating cells. Finally, the ability to isolate live LRCC and LRC has implications beyond cancer; it may provide novel insights into normal adult tissue stem cells, tissue regeneration, and tissue degeneration into cancer.
Summary
LRCs are thought to be tissue stem cells. GI cancers are derived from tissues containing LRCs. We show, in live cells, that various GI cancers contain LRCCs undergoing ACD and exhibit stem cells and pluripotency gene expression profiles. Importantly, LRCCs have greater tumor-initiating capacity than non-LRCC. We propose that LRCC might represent a novel population of stem-like cancer cells.
Supplementary Material
Acknowledgment
This study was supported by the intramural grant provided by the NIH/National Cancer Institute.
Footnotes
Author contributions: H.-W.X: concept and design, collection and assembly of data, data analysis and interpretation, and manuscript writing; D.M.H., J.E.M., C.M.A., A.J.A., G.W.W., and S.H.G.: collection and assembly of data; T.K. and S.R.: manuscript writing; S.S.T.: data analysis and interpretation and manuscript writing; I.A: concept and design, data analysis and interpretation, manuscript writing, and final approval of manuscript. H.-W.X. and D.M.H. contributed equally to this article.
Disclosure of Potential Conflicts of Interest The authors declare no potential conflicts of interests.
References
- 1.Lark KG, Consigli RA, Minocha HC. Segregation of sister chromatids in mammalian cells. Science. 1966;154:1202–1205. doi: 10.1126/science.154.3753.1202. [DOI] [PubMed] [Google Scholar]
- 2.Potten CS, Hume WJ, Reid P, et al. The segregation of DNA in epithelial stem cells. Cell. 1978;15:899–906. doi: 10.1016/0092-8674(78)90274-x. [DOI] [PubMed] [Google Scholar]
- 3.Cotsarelis G, Sun TT, Lavker RM. Label-retaining cells reside in the bulge area of pilosebaceous unit: Implications for follicular stem cells, hair cycle, and skin carcinogenesis. Cell. 1990;61:1329–1337. doi: 10.1016/0092-8674(90)90696-c. [DOI] [PubMed] [Google Scholar]
- 4.Smith GH. Label-retaining epithelial cells in mouse mammary gland divide asymmetrically and retain their template DNA strands. Development. 2005;132:681–687. doi: 10.1242/dev.01609. [DOI] [PubMed] [Google Scholar]
- 5.Shinin V, Gayraud-Morel B, Gomes D, et al. Asymmetric division and cosegregation of template DNA strands in adult muscle satellite cells. Nat Cell Biol. 2006;8:677–687. doi: 10.1038/ncb1425. [DOI] [PubMed] [Google Scholar]
- 6.Karpowicz P, Morshead C, Kam A, et al. Support for the immortal strand hypothesis: Neural stem cells partition DNA asymmetrically in vitro. J Cell Biol. 2005;170:721–732. doi: 10.1083/jcb.200502073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li F, Lu L, Lu J. Identification and location of label retaining cells in mouse liver. J Gastroenterol. 2010;45:113–121. doi: 10.1007/s00535-009-0139-2. [DOI] [PubMed] [Google Scholar]
- 8.Zabierowski SE, Herlyn M. Melanoma stem cells: The dark seed of melanoma. J Clin Oncol. 2008;26:2890–2894. doi: 10.1200/JCO.2007.15.5465. [DOI] [PubMed] [Google Scholar]
- 9.Moore N, Lyle S. Quiescent, slow-cycling stem cell populations in cancer: A review of the evidence and discussion of significance. J Oncol. 2011;2011 doi: 10.1155/2011/396076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Potten CS, Owen G, Booth D. Intestinal stem cells protect their genome by selective segregation of template DNA strands. J Cell Sci. 2002;115:2381–2388. doi: 10.1242/jcs.115.11.2381. [DOI] [PubMed] [Google Scholar]
- 11.Cairns J. Mutation selection and the natural history of cancer. Nature. 1975;255:197–200. doi: 10.1038/255197a0. [DOI] [PubMed] [Google Scholar]
- 12.Pine SR, Ryan BM, Varticovski L, et al. Microenvironmental modulation of asymmetric cell division in human lung cancer cells. Proc Natl Acad Sci USA. 2010;107:2195–2200. doi: 10.1073/pnas.0909390107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Conboy MJ, Karasov AO, Rando TA. High incidence of non-random template strand segregation and asymmetric fate determination in dividing stem cells and their progeny. PLoS Biol. 2007;5:e102. doi: 10.1371/journal.pbio.0050102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rambhatla L, Ram-Mohan S, Cheng JJ, et al. Immortal DNA strand cosegregation requires p53/IMPDH-dependent asymmetric self-renewal associated with adult stem cells. Cancer Res. 2005;65:3155–3161. doi: 10.1158/0008-5472.CAN-04-3161. [DOI] [PubMed] [Google Scholar]
- 15.Walters K. Colonic stem cell data are consistent with the immortal model of stem cell division under non-random strand segregation. Cell Prolif. 2009;42:339–347. doi: 10.1111/j.1365-2184.2009.00600.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Falconer E, Chavez EA, Henderson A, et al. Identification of sister chromatids by DNA template strand sequences. Nature. 2010;463:93–97. doi: 10.1038/nature08644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kiel MJ, He S, Ashkenazi R, et al. Haematopoietic stem cells do not asymmetrically segregate chromosomes or retain BrdU. Nature. 2007;449:238–242. doi: 10.1038/nature06115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lansdorp PM. Immortal strands? Give me a break. Cell. 2007;129:1244–1247. doi: 10.1016/j.cell.2007.06.017. [DOI] [PubMed] [Google Scholar]
- 19.Karpowicz P, Pellikka M, Chea E, et al. The germline stem cells of Drosophila melanogaster partition DNA non-randomly. Eur J Cell Biol. 2009;88:397–408. doi: 10.1016/j.ejcb.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 20.Fei JF, Huttner WB. Nonselective sister chromatid segregation in mouse embryonic neocortical precursor cells. Cereb Cortex. 2009;19(suppl 1):i49–i54. doi: 10.1093/cercor/bhp043. [DOI] [PubMed] [Google Scholar]
- 21.Sotiropoulou PA, Candi A, Blanpain C. The majority of multipotent epidermal stem cells do not protect their genome by asymmetrical chromosome segregation. Stem Cells. 2008;26:2964–2973. doi: 10.1634/stemcells.2008-0634. [DOI] [PubMed] [Google Scholar]
- 22.Yadlapalli S, Cheng J, Yamashita YM. Drosophila male germline stem cells do not asymmetrically segregate chromosome strands. J Cell Sci. 2011;124:933–939. doi: 10.1242/jcs.079798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schepers AG, Vries R, van den Born M, et al. Lgr5 intestinal stem cells have high telomerase activity and randomly segregate their chromosomes. EMBO J. 2011;30:1104–1109. doi: 10.1038/emboj.2011.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lapidot T, Sirard C, Vormoor J, et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367:645–648. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- 25.Boiko AD, Razorenova OV, van de Rijn M, et al. Human melanoma-initiating cells express neural crest nerve growth factor receptor CD271. Nature. 2010;466:133–137. doi: 10.1038/nature09161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: Accumulating evidence and unresolved questions. Nat Rev Cancer. 2008;8:755–768. doi: 10.1038/nrc2499. [DOI] [PubMed] [Google Scholar]
- 27.Al-Hajj M, Wicha MS, Benito-Hernandez A, et al. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma S, Chan K-W, Hu L, et al. Identification and characterization of tumorigenic liver cancer stem/progenitor cells. Gastroenterology. 2007;132:2542–2556. doi: 10.1053/j.gastro.2007.04.025. [DOI] [PubMed] [Google Scholar]
- 29.O'Brien CA, Pollett A, Gallinger S, et al. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445:106–110. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- 30.Singh SK, Hawkins C, Clarke ID, et al. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 31.Goodell MA, Brose K, Paradis G, et al. Isolation and functional properties of murine hematopoietic stem cells that are replicating in vivo. J Exp Med. 1996;183:1797–1806. doi: 10.1084/jem.183.4.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li C, Heidt DG, Dalerba P, et al. Identification of pancreatic cancer stem cells. Cancer Res. 2007;67:1030–1037. doi: 10.1158/0008-5472.CAN-06-2030. [DOI] [PubMed] [Google Scholar]
- 33.Sohn J, Natale J, Chew LJ, et al. Identification of Sox17 as a transcription factor that regulates oligodendrocyte development. J Neurosci. 2006;26:9722–9735. doi: 10.1523/JNEUROSCI.1716-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sinner D, Rankin S, Lee M, et al. Sox17 and beta-catenin cooperate to regulate the transcription of endodermal genes. Development. 2004;131:3069–3080. doi: 10.1242/dev.01176. [DOI] [PubMed] [Google Scholar]
- 35.Park IH, Zhao R, West JA, et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 36.Yu J, Vodyanik MA, Smuga-Otto K, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 37.Pera MF, Tam PP. Extrinsic regulation of pluripotent stem cells. Nature. 2010;465:713–720. doi: 10.1038/nature09228. [DOI] [PubMed] [Google Scholar]
- 38.Balciunaite G, Keller MP, Balciunaite E, et al. Wnt glycoproteins regulate the expression of FoxN1, the gene defective in nude mice. Nature Immunol. 2002;3:1102–1108. doi: 10.1038/ni850. [DOI] [PubMed] [Google Scholar]
- 39.Ewart JL, Cohen MF, Meyer RA, et al. Heart and neural tube defects in transgenic mice overexpressing the Cx43 gap junction gene. Development. 1997;124:1281–1292. doi: 10.1242/dev.124.7.1281. [DOI] [PubMed] [Google Scholar]
- 40.Jang J, Choi YI, Choi J, et al. Notch1 confers thymocytes a resistance to GC-induced apoptosis through Deltex1 by blocking the recruitment of p300 to the SRG3 promoter. Cell Death Differ. 2006;13:1495–1505. doi: 10.1038/sj.cdd.4401827. [DOI] [PubMed] [Google Scholar]
- 41.Izon DJ, Aster JC, He Y, et al. Deltex1 redirects lymphoid progenitors to the B cell lineage by antagonizing Notch1. Immunity. 2002;16:231–243. doi: 10.1016/s1074-7613(02)00271-6. [DOI] [PubMed] [Google Scholar]
- 42.Katoh M, Katoh M. Notch signaling in gastrointestinal tract (review) Int J Oncol. 2007;30:247–251. [PubMed] [Google Scholar]
- 43.Sumitomo M, Shen R, Walburg M, et al. Neutral endopeptidase inhibits prostate cancer cell migration by blocking focal adhesion kinase signaling. J Clin Invest. 2000;106:1399–1407. doi: 10.1172/JCI10536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kanitakis J, Narvaez D, Claudy A. Differential expression of the CD10 antigen (neutral endopeptidase) in primary versus metastatic malignant melanomas of the skin. Melanoma Res. 2002;12:241–244. doi: 10.1097/00008390-200206000-00007. [DOI] [PubMed] [Google Scholar]
- 45.Brinkmann MM, Pietrek M, Dittrich-Breiholz O, et al. Modulation of host gene expression by the K15 protein of Kaposi's sarcoma-associated herpesvirus. J Virol. 2007;81:42–58. doi: 10.1128/JVI.00648-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Astigiano S, Damonte P, Barbieri O. Inhibition of ductal morphogenesis in the mammary gland of WAP-fgf4 transgenic mice. Anat Embryol. 2003;206:471–478. doi: 10.1007/s00429-003-0317-6. [DOI] [PubMed] [Google Scholar]
- 47.Maerz WJ, Baselga J, Reuter VE, et al. FGF4 dissociates anti-tumorigenic from differentiation signals of retinoic acid in human embryonal carcinomas. Oncogene. 1998;17:761–767. doi: 10.1038/sj.onc.1201992. [DOI] [PubMed] [Google Scholar]
- 48.Nishimoto M, Fukushima A, Okuda A, et al. The gene for the embryonic stem cell coactivator UTF1 carries a regulatory element which selectively interacts with a complex composed of Oct-3/4 and Sox-2. Mol Cell Biol. 1999;19:5453–5465. doi: 10.1128/mcb.19.8.5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yuan H, Corbi N, Basilico C, et al. Developmental-specific activity of the FGF-4 enhancer requires the synergistic action of Sox2 and Oct-3. Genes Dev. 1995;9:2635–2645. doi: 10.1101/gad.9.21.2635. [DOI] [PubMed] [Google Scholar]
- 50.Lee HX, Mendes FA, Plouhinec JL, et al. Enzymatic regulation of pattern: BMP4 binds CUB domains of Tolloids and inhibits proteinase activity. Genes Dev. 2009;23:2551–2562. doi: 10.1101/gad.1839309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zheng H, Fu G, Dai T, et al. Migration of endothelial progenitor cells mediated by stromal cell-derived factor-1alpha/CXCR4 via PI3K/Akt/eNOS signal transduction pathway. J Cardiovasc Pharmacol. 2007;50:274–280. doi: 10.1097/FJC.0b013e318093ec8f. [DOI] [PubMed] [Google Scholar]
- 52.Barker N, van Es JH, Kuipers J, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 53.Guo Y, Chen Y, Ito H, et al. Identification and characterization of lin-28 homolog B (LIN28B) in human hepatocellular carcinoma. Gene. 2006;384:51–61. doi: 10.1016/j.gene.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 54.Richards M, Tan SP, Tan JH, et al. The transcriptome profile of human embryonic stem cells as defined by SAGE. Stem Cells. 2004;22:51–64. doi: 10.1634/stemcells.22-1-51. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.