Figure 5.
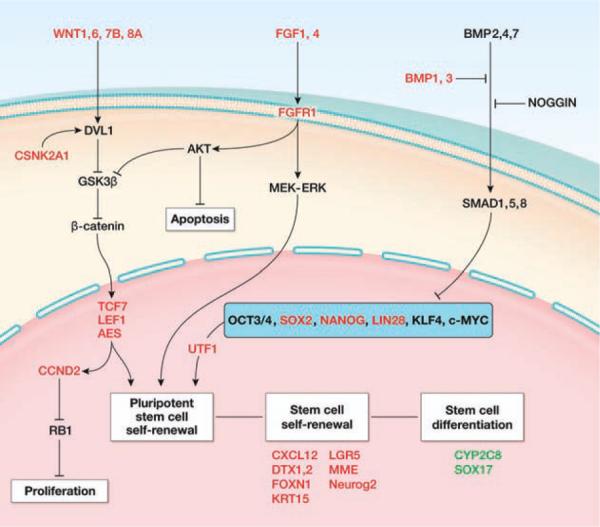
Stem cell pathways analysis for label-retaining cancer cell (LRCC). Using our gene expression data and the ingenuity pathway analysis platform, we show that LRCC upregulates known stem cells, pluripotency, and Wnt pathway genes and downregulates genes associated with gastrointestinal differentiation and apoptosis suggesting a stem cell gene expression profile. Abbreviations: AES, amino-terminal enhancer of split; AKT, v-akt murine thymoma viral oncogene homolog; BMP, bone morphogenetic protein; CCND, cyclin D; CSNK, casein kinase; CXCL, chemokine (C-X-C motif) ligand; CYP, cytochrome P450; DTX, deltex homolog; DVL, dishevelled; FGF, fibroblast growth factor; FOXN, fork-head box N; GSK, glycogen synthase kinase; KLF, Kruppel-like factor; KRT, keratin; LEF, lymphoid enhancer-binding factor; LGR, leucine-rich repeat containing G protein-coupled receptor; LIN28, lin-28 homolog; MEK-ERK, mitogen-activated protein kinase kinase-MAP kinase; MME, membrane metallo-endopeptidase; MYC, v-myc myelocytomatosis viral oncogene homolog; NANOG, nanog homeobox; NEUROG, neurogenin; NOGGIN, noggin; OCT, octamer-binding protein; RB, retinoblastoma; SMAD, mothers against DPP homolog; SOX, SRY (sex determining region Y)-box; TCF, T-cell specific transcription factor; UTF, undifferentiated embryonic cell transcription factor; WNT, Wingless-type MMTV integration site family.
