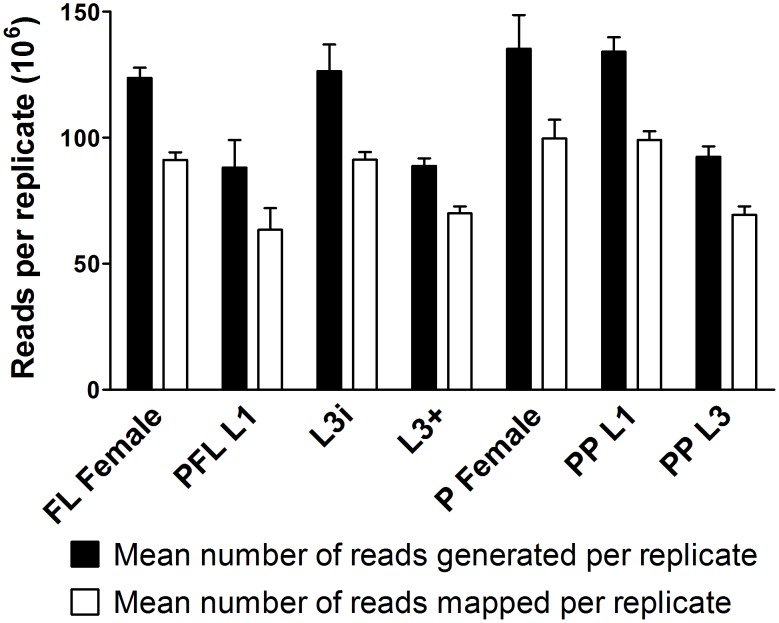
Figure 3. S. stercoralis RNAseq mean library sizes and number of reads aligning to the genome.
A total of 21 libraries were derived from polyadenylated RNA and sequenced from seven developmental stages, each in biological triplicate. Paired-end 100 base-pair (bp) reads were generated from the following developmental stages: free-living females (FL Female), post-free-living first-stage larvae (PFL L1), infectious third-stage larvae (L3i), in vivo activated third-stage larvae (L3+), parasitic females (P Female), predominantly (>95%) heterogonically developing post-parasitic first-stage larvae (PP L1), and post-parasitic approximately third-stage larvae heterogonically developing to free-living adults and enriched for females (PP L3). The mean number of reads generated per replicate refers to the mean number of 100 bp reads sequenced (black bars) per biological replicate from each developmental stage. The mean number of mapped reads per replicate refers to the mean number of 100 bp reads aligned to S. stercoralis genomic contigs using TopHat (white bars) per biological replicate from each developmental stage. Error bars represent +1 standard deviation.