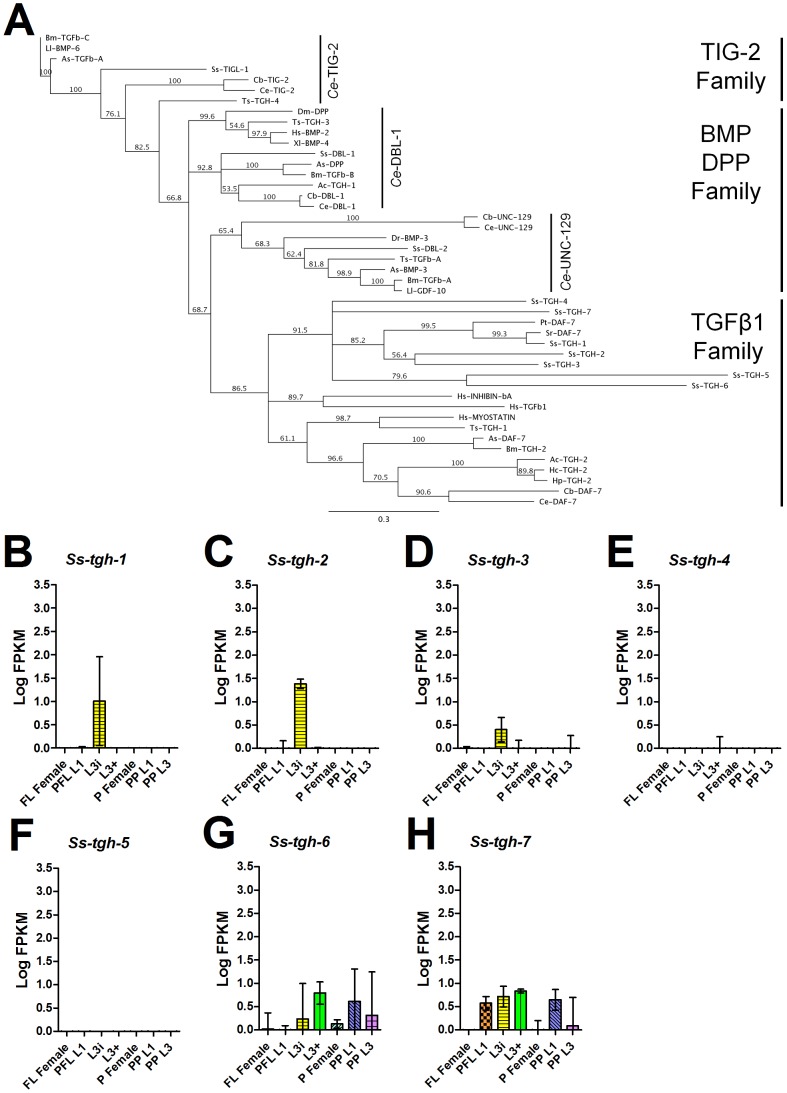
Figure 6. Phylogenetic analysis and temporal regulation of S. stercoralis TGFβ ligands.
(A) Phylogenetic analysis of the transforming growth factor β (TGFβ) super-family ligands was performed; nematode TGFβ ligands resolved into three main families that share the same cysteine architecture. Ss-TIGL-1 groups with the Ce-TIG-2-like family; Ss-DBL-1 and Ss-DBL-2 group with the D. melanogaster decapentaplegic (DPP) and vertebrate bone morphogenetic protein (BMP) family; and Ss-TGH-1 through -7 group with the human TGFβ1 family that also includes Ce-DAF-7. A Clustal W alignment of the TGFβ ligands truncated at the first conserved cysteine was used to construct the neighbor-joining tree with 100 iterations of boot-strapping. Abbreviations: Ancylostoma caninum (Ac), Ascaris suum (As), Brugia malayi (Bm), Caenorhabditis briggsae (Cb), Caenorhabditis elegans (Ce), Danio rerio (Dr), Drosophila melanogaster (Dm), Haemonchus contortus (Hc), Heligmosomoides polygyrus (Hp), Homo sapiens (Hs), Loa loa (Ll), Parastrongyloides trichosuri (Pt), Strongyloides ratti (Sr), Strongyloides stercoralis (Ss), Trichinella spiralis (Ts), and Xenopus laevis (Xl). The scale bar represents substitutions per position. Accession numbers are listed in Data S4. (B–H) Transcript abundances were determined for the coding region of seven S. stercoralis genes, Ss-tgh-1 through -7, encoding putative TGFβ ligands similar to Ce-DAF-7 in seven developmental stages: free-living females (FL Female), post-free-living first-stage larvae (PFL L1), infectious third-stage larvae (L3i), in vivo activated third-stage larvae (L3+), parasitic females (P Female), post-parasitic first-stage larvae (PP L1), and post-parasitic third-stage larvae (PP L3). Transcript abundances were calculated as fragments per kilobase of coding exon per million mapped reads (FPKM) and log transformed. Error bars represent 95% confidence intervals. The y-axes were scaled from 0 to 3.5 to aid comparison between genes.