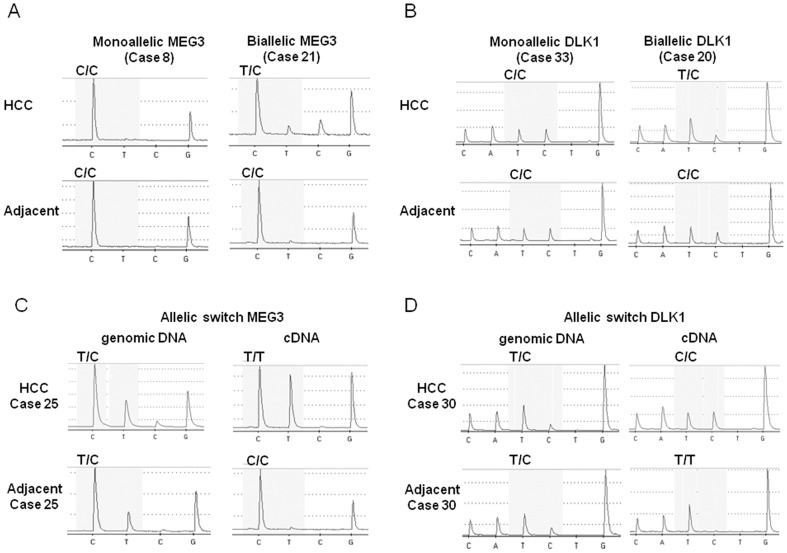
Figure 6. Allele-specific expression analysis of the DLK1-MEG3 locus in human HCC.
Quantitative SNP analysis was performed using pyrosequencing at a C/G polymorphism in exon 3, at SNP Rs8013873 for MEG3 and SNP Rs1802710 for DLK1. In tumour specimens showing genomic DNA polymorphisms, further SNP analysis was performed with the cDNAs from the same sample as well as with genomic and cDNA from the corresponding adjacent non-cancerous liver tissues. (A) Pyrosequencing analysis of the cDNA at SNP Rs8013873 shows mono-allelic MEG3 expression in HCC specimen #8 and bi-allelic expression in HCC specimen #21. (B) Pyrosequencing analysis of the cDNA at SNP Rs1802710 shows mono-allelic DLK1 expression in HCC specimen #33 and bi-allelic expression in HCC specimen #20. (C) MEG3 allelic switching is shown in case #25. Informative polymorphisms are found in the genomic DNAs of both tumour and the adjacent non-cancerous tissues and T/T expression in cDNA tumour but C/C expression in cDNA adjacent liver tissues. (D) DLK1 allelic switching is shown in case #30 with informative polymorphisms in the genomic DNAs of both tumour and the adjacent non-cancerous tissues and C/C expression in tumour cDNA but T/T expression in cDNA from the adjacent liver tissues. The confirmation of all genotyping results by Sanger sequencing is shown in Figure S5.