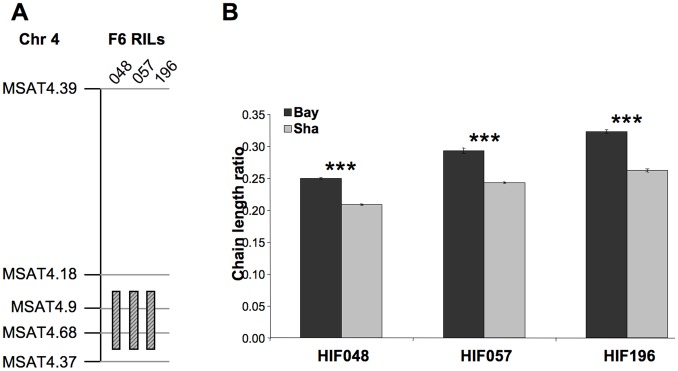
Figure 1. Confirmation of the CLR.2 QTL with heterogeneous inbred families (HIFs).
A, F6 recombinant inbred lines (RILs) from the Bay-0 x Shahdara cross showing residual heterozygosity in the region of the CLR.2 QTL. Numbers on the top designate the RIL, with the hatched black and grey bar beneath indicating the region still segregating. Recombination breakpoints delimiting heterozygous regions are arbitrarily depicted in the middle of the marker interval. The vertical black line represents chromosome 4 with markers indicated on the left. Marker positions and identity can be found at www.inra.fr/vast/in the MSAT database. B, Comparison of CLR for HIFs fixed for the Bay (black) or the Sha (grey) allele at the segregating region. CLR was determined from 20 seeds per plants. Bars represent SE values (n = 8, 4 repetitions were done on 2 plants). Significance in t-test, ***p<10−8.