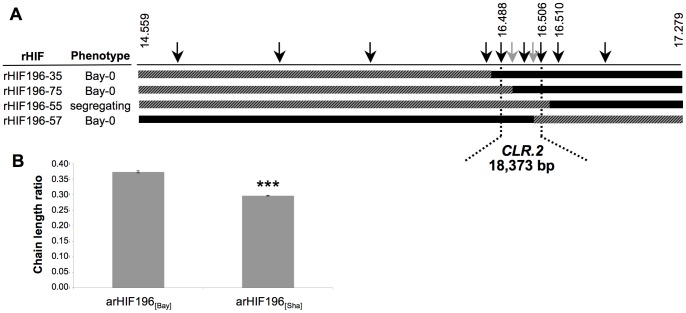
Figure 2. Fine mapping of CLR.2.
A, The genotype of the most informative recombinants (rHIFs) used to delineate the final 18,373-bp candidate interval are represented with horizontal bars (black for Bay allele, grey for Sha allele, hatched black/grey for heterozygous). Vertical black arrows above represent markers used for genotyping between 14.559 Mb and 17.279 Mb on chromosome 4. Numbers indicate physical position in Mb (TAIR8). Grey arrows mark the most informative recombination breakpoints. B, Chain length ratio of arHIF196[Bay] and arHIF196[Sha] (Materials and Methods) was compared. CLR was determined on 100 seeds from 4 independent plants for each arHIF. Bars represent SE values (n = 4). Significance in t-test, ***p<10−6.