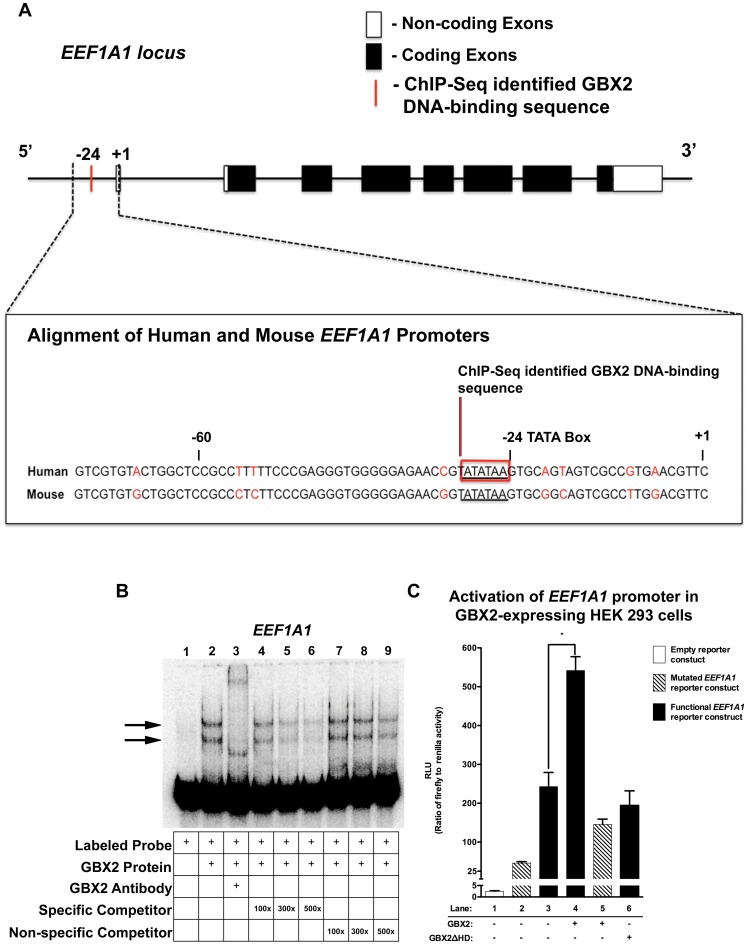
Figure 6. GBX2 binds to and functionally interacts within the EEF1A1 core promoter.
(A) EEF1A1 locus depicting non-coding exons (white boxes), coding exons (black boxes), and the ChIP-Seq identified location of the GBX2 DNA-binding sequence (red bar). Alignment of the human and mouse ChIP-Seq identified EEF1A1 promoter region using sequences obtained from Ensembl [61], EEF1A1 TATA box (underlined sequence) and GBX2 DNA-binding sequence (red box). (B) Gel-shift analysis for identified GBX2 target EEF1A1. A reduction in the mobility of [ÿ -32P] ATP labeled EEF1A1 100-mer probe is observed with the addition of GBX2 (black arrows). A supershift is observed in lane 3 with the addition of anti-GBX2. Addition of identical EEF1A1 100-mer unlabeled specific competitor probes at 100x, 300x, and 500x molar concentrations in lanes 4–6. Addition of EEF1A1 45-mer unlabeled non-specific competitor probes, omitting the GBX2 DNA-binding sequence in lanes 7–9. (C) EEF1A1 promoter luciferase reporter assay. HEK 293 cells were transiently transfected with either the empty pGL4.10[luc2] vector (white bar), the pGL4.10[luc2] vector containing the functional EEF1A1 promoter sequence and the TATA box (TATATAA; black bars), the pGL4.10[luc2] vector containing the mutated EEF1A1 promoter sequence with a mutated TATA box (TATATAA changed to GCGCGCC; striped bars), and the pGL4.70[hRluc] Renilla vector. Substantial luciferase activity was observed in cells transfected with the pGL4.10[luc2] vector containing the functional EEF1A1 promoter compared to the empty pGL4.10[luc2] reporter construct (compare lane 3 to lane 1). Maximal luciferase activation is observed upon the addition of GBX2 in cells with the pGL4.10[luc2] vector containing the functional EEF1A1 promoter sequence (compare lane 4 to lane 3), and activation is reduced in GBX2ΔHD (lane 6) cells and cells expressing GBX2 and the pGL4.10[luc2] mutated EEF1A1 reporter construct (compare lane 4 to lane 5). Luciferase activities were normalized to Renilla luciferase activities. * P = 0.0047 (two-tailed P value).