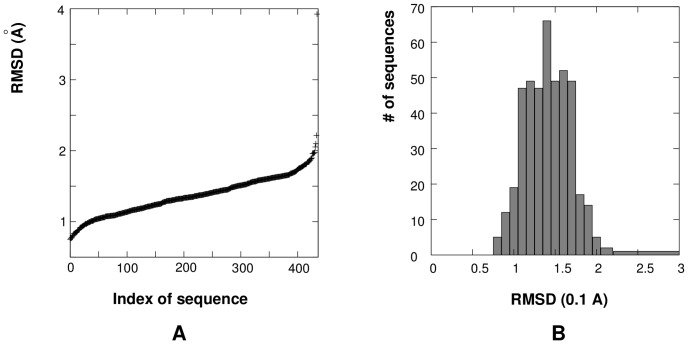
Figure 8. ( ) The structural differences (RMSD values) between PDB structures and diamond lattice-represented structures for the RNA loops/junctions within the Leontis dataset.
) The structural differences (RMSD values) between PDB structures and diamond lattice-represented structures for the RNA loops/junctions within the Leontis dataset.
The x-axis represents the index of RNA loops/junctions in the Leontis dataset. (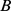 ) The number of RNA loops/junctions within each RMSD-value bin (0.1 Å). The mean and standard deviation of RMSD values for the 435 RNA loops/junctions are 1.37 Å and 0.29 Å, respectively.
) The number of RNA loops/junctions within each RMSD-value bin (0.1 Å). The mean and standard deviation of RMSD values for the 435 RNA loops/junctions are 1.37 Å and 0.29 Å, respectively.