Table 4. The numbers of sequences of the successfully predicted loop/junction structures for (I) all 8452 RNA loops/junctions in TEST-I (II) the 7459 RNA loops and junctions in TEST-II and (III) the 1119 RNA loops and junctions in TEST-III.
| TEST-I | ||||
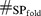 (A) (A) |
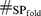 (B) (B) |
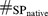 (A) (A) |
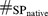 (B) (B) |
|
| Top-1 | 7364 | 7369 | 6992 | 7102 |
| Top-2 | 7686 | 7686 | 7411 | 7500 |
| Top-3 | 7796 | 7862 | 7715 | 7665 |
| Top-4 | 7956 | 7969 | 7798 | 7844 |
| Top-5 | 8006 | 8007 | 7857 | 7878 |
TOTAL-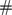
|
8452 | |||
For each dataset, the numbers in columns “A” are calculated from 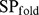 (“
(“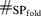 ”, “true” potential parameters) and
”, “true” potential parameters) and 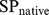 (“
(“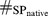 ”, “extracted” potential parameters), obtained from the RNA09 database, and the numbers in columns “B” are calculated from
”, “extracted” potential parameters), obtained from the RNA09 database, and the numbers in columns “B” are calculated from 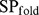 (“
(“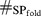 ”) and
”) and 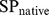 (“
(“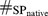 ”), obtained from the Leontis' database respectively. The “Top-
”), obtained from the Leontis' database respectively. The “Top-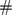 ” in the first column means that the “correct” structure is in the top
” in the first column means that the “correct” structure is in the top 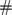 lowest-potential conformations.
lowest-potential conformations.
