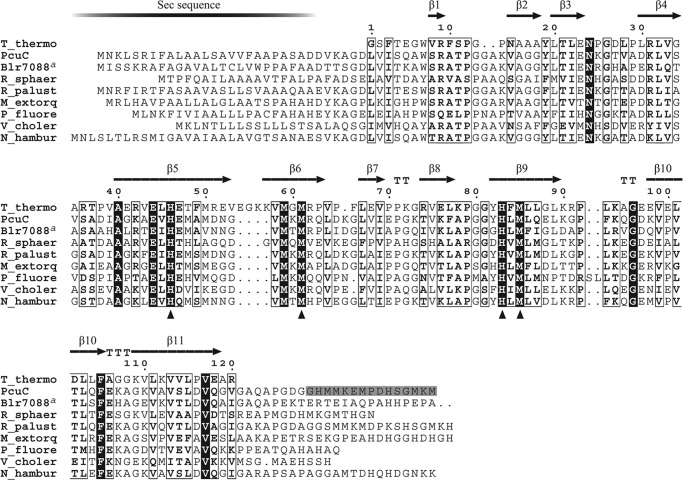
FIGURE 3.
Multiple alignment of PCuAC amino acidic sequences. The secondary structure of T. thermophilus PCuAC extracted from its crystal structure (Protein Data Bank code 2K6W) is shown above the alignment. The numbering refers to the T. thermophilus PCuAC sequence. B. japonicum PcuC and Blr7088, R. sphaeroides 2.4.1 RSP_2017, Rhodopseudomonas palustris HaA2 RPB_2549, Methylobacterium extorquens PA1 Mext_1379, Pseudomonas fluorescens Pf0-1Pfl01_0598, Vibrio cholerae HC-02A1 VCHC02A1_2964, and Nitrobacter hamburgensis X14 Nham_2200 sequences are included in the alignment. Similarity and identity columns are shown as empty boxes and white on black type, respectively. Copper-binding residues of TtPCuAC are marked with triangles; the corresponding residues of PcuC are His79, Met90, His113, and Met115. The gray-shaded amino acids at the C-terminal end of the PcuC sequence are those that were removed to test if they bind copper. a, the C-terminal 118-amino acid domain of Blr7088 was omitted in the alignment.